1NFK
 
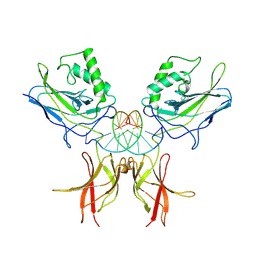 | | STRUCTURE OF THE NUCLEAR FACTOR KAPPA-B (NF-KB) P50 HOMODIMER | | Descriptor: | DNA (5'-D(*TP*GP*GP*GP*AP*AP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Ghosh, G, Van Duyne, G, Ghosh, S, Sigler, P.B. | | Deposit date: | 1995-02-28 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of NF-kappa B p50 homodimer bound to a kappa B site.
Nature, 373, 1995
|
|
1NFN
 
 | | APOLIPOPROTEIN E3 (APOE3) | | Descriptor: | APOLIPOPROTEIN E3 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFO
 
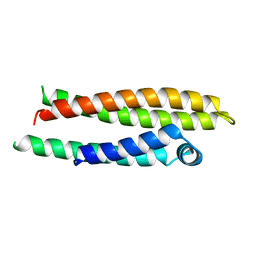 | | APOLIPOPROTEIN E2 (APOE2, D154A MUTATION) | | Descriptor: | APOLIPOPROTEIN E2 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFP
 
 | |
1NFQ
 
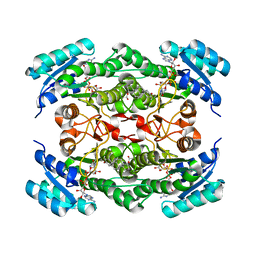 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFS
 
 | |
1NFT
 
 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1NFU
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR132747 | | Descriptor: | 3-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1NFV
 
 | | X-ray structure of Desulfovibrio desulfuricans bacterioferritin: the diiron centre in different catalytic states (as-isolated structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, 3-HYDROXYPYRUVIC ACID, FE (III) ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NFW
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR209685 | | Descriptor: | 4-{[(E)-2-(5-CHLOROTHIEN-2-YL)VINYL]SULFONYL}-1-(1H-PYRROLO[3,2-C]PYRIDIN-2-YLMETHYL)PIPERAZIN-2-ONE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors:
preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1NFX
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR208944 | | Descriptor: | 4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-1-{[1-(2-HYDROXYETHYL)-1H-PYRROLO[3,2-C]PYRIDIN-2-YL]METHYL}PIPERAZIN-2-ONE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors:
preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1NFY
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR200095 | | Descriptor: | 4-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, Coagulation factor XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
1NFZ
 
 | |
1NG0
 
 | |
1NG1
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-04-30 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
1NG2
 
 | | Structure of autoinhibited p47phox | | Descriptor: | Neutrophil cytosolic factor 1 | | Authors: | Groemping, Y, Lapouge, K, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2002-12-16 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of phosphorylation-induced activation of the NADPH oxidase
Cell(Cambridge,Mass.), 113, 2003
|
|
1NG3
 
 | | Complex of ThiO (glycine oxidase) with acetyl-glycine | | Descriptor: | ACETYLAMINO-ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1NG4
 
 | | Structure of ThiO (glycine oxidase) from Bacillus subtilis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, HYDROGEN PEROXIDE, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1NG5
 
 | |
1NG6
 
 | | Structure of Cytosolic Protein of Unknown Function YqeY from Bacillus subtilis | | Descriptor: | Hypothetical protein yqeY | | Authors: | Zhang, R, Dementiva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-16 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4A crystal structure of hypothetical cytosolic protein
YQEY
To be Published
|
|
1NG7
 
 | |
1NG8
 
 | |
1NG9
 
 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2002-12-17 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
1NGA
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
