8J6M
 
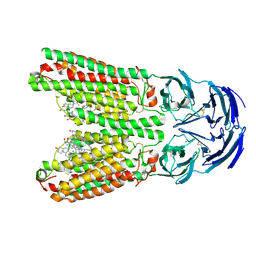 | | SIDT1 protein | | Descriptor: | CHOLESTEROL, Green fluorescent protein,SID1 transmembrane family member 1, OLEIC ACID, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8K75
 
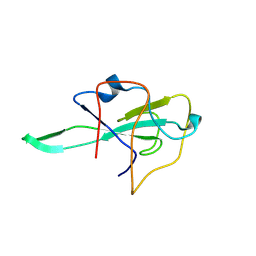 | |
8Y80
 
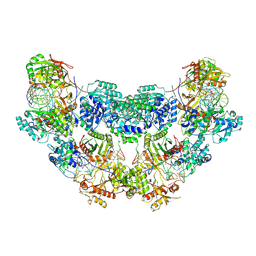 | | Cryo-EM structure of the tetrameric SPARSA gRNA-ssDNA complex | | Descriptor: | DNA (25-mer), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, J.T, Cui, N, Wei, X.Y, Jia, N. | | Deposit date: | 2024-02-05 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
8Y7Z
 
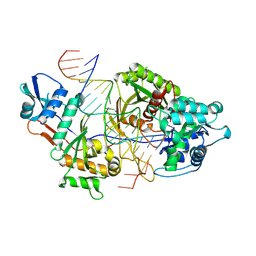 | | Cryo-EM structure of the monomeric SPARSA gRNA-ssDNA complex | | Descriptor: | DNA (25-mer), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, J.T, Cui, N, Wei, X.Y, Jia, N. | | Deposit date: | 2024-02-05 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
8Y82
 
 | | Cryo-EM structure of the tetrameric SPARSA gRNA-ssDNA-NAD+ complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (25-mer), MAGNESIUM ION, ... | | Authors: | Zhang, J.T, Cui, N, Wei, X.Y, Jia, N. | | Deposit date: | 2024-02-05 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tetramerization-dependent activation of the Sir2-associated short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
8WQK
 
 | |
8K34
 
 | | Cryo-EM structure of SPARTA gRNA binary complex | | Descriptor: | MAGNESIUM ION, Piwi domain-containing protein, RNA (5'-R(P*AP*AP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Zhang, J.T, Jia, N. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Target ssDNA activates the NADase activity of prokaryotic SPARTA immune system.
Nat.Chem.Biol., 20, 2024
|
|
8WYD
 
 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY9
 
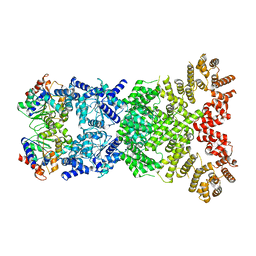 | |
8WYE
 
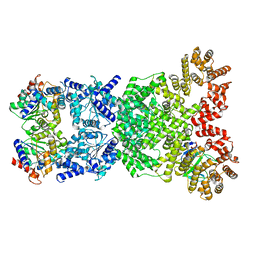 | | Cryo-EM structure of DSR2-DSAD1 (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYA
 
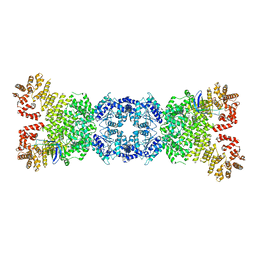 | | Cryo-EM structure of DSR2-tube complex | | Descriptor: | Bacillus phage SPbeta tube protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYB
 
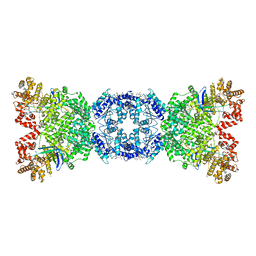 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ complex | | Descriptor: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY8
 
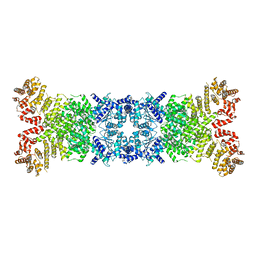 | | Cryo-EM structure of DSR2 apo complex | | Descriptor: | SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYF
 
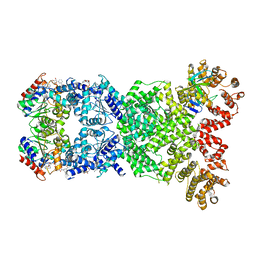 | | Cryo-EM structure of DSR2-DSAD1-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYC
 
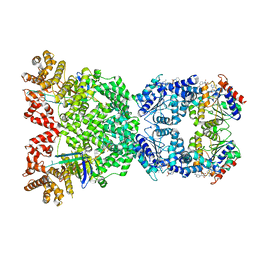 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8IFM
 
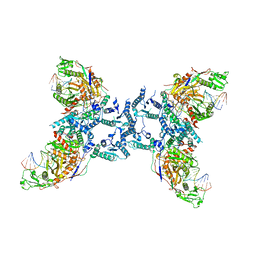 | |
8IFL
 
 | |
8IFK
 
 | |
7XM9
 
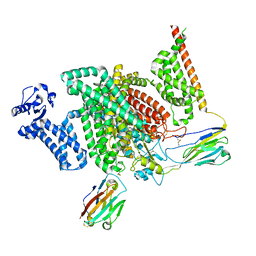 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XMF
 
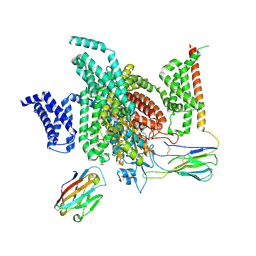 | | Cryo-EM structure of human NaV1.7/beta1/beta2-Nav1.7-IN2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[4-[3-(4-fluoranyl-2-methyl-phenoxy)azetidin-1-yl]pyrimidin-2-yl]amino]-~{N}-methyl-benzamide, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7EEL
 
 | | Cyanophage Pam1 capsid asymmetric unit | | Descriptor: | Cement (decoration) proteins, Major capsid proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEA
 
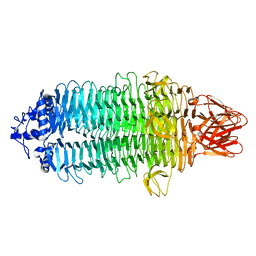 | |
7EEQ
 
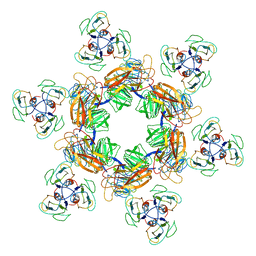 | | Cyanophage Pam1 tail machine | | Descriptor: | Needle head proteins, Tailspike head-binding domain | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEP
 
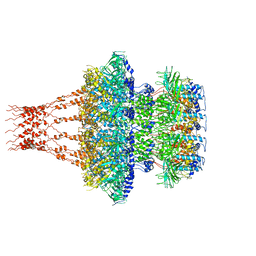 | | Cyanophage Pam1 portal-adaptor complex | | Descriptor: | Pam1 adaptor proteins, Pam1 portal proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7XF0
 
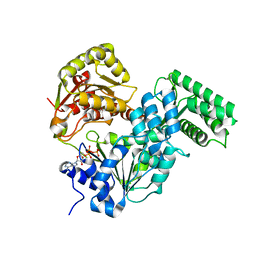 | | Crystal strucutre of CasDinG in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CasDinG | | Authors: | Zhang, J.T, Cui, N, Liu, Y.R, Huang, H.D, Jia, N. | | Deposit date: | 2022-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
