7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7XD2
 
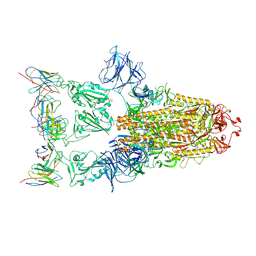 | |
7CYP
 
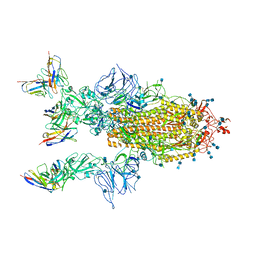 | | Complex of SARS-CoV-2 spike trimer with its neutralizing antibody HB27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of HB27, ... | | Authors: | Wang, X, Zhu, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-06-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
5B7I
 
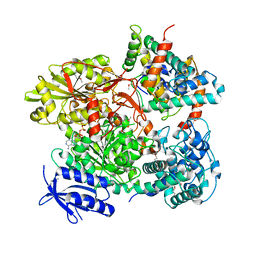 | | Cas3-AcrF3 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2016-07-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3
Nat.Struct.Mol.Biol., 23, 2016
|
|
7LGI
 
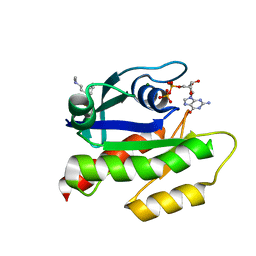 | | The haddock model of GDP KRas in complex with promazine using chemical shift perturbations and intermolecular NOEs | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
8HKN
 
 | |
8HLJ
 
 | |
8HKO
 
 | |
8HLQ
 
 | |
2GAS
 
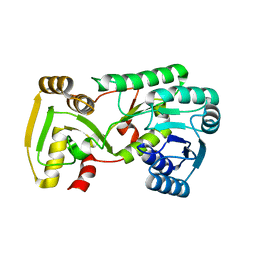 | | Crystal Structure of Isoflavone Reductase | | Descriptor: | isoflavone reductase | | Authors: | Wang, X, He, X, Lin, J, Shao, H, Chang, Z, Dixon, R.A. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Isoflavone Reductase from Alfalfa (Medicago sativa L.)
J.Mol.Biol., 358, 2006
|
|
7VFX
 
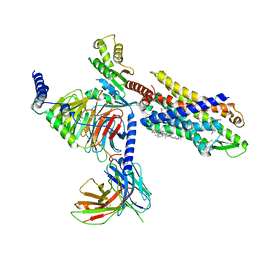 | | The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-21 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
6V5L
 
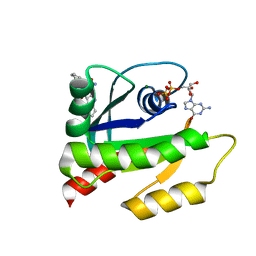 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
7MQU
 
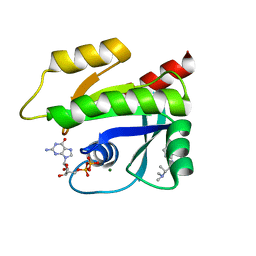 | | The haddock model of GDP KRas in complex with promethazine using NMR chemical shift perturbations | | Descriptor: | (2R)-N,N-dimethyl-1-(10H-phenothiazin-10-yl)propan-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.P. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
5CO5
 
 | |
8WLZ
 
 | | Cryo-EM structure of the WIV1 S-hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-01 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1
J.Virol., 2024
|
|
8WLY
 
 | | Cryo-EM structure of bat WIV1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-01 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1
J.Virol., 2024
|
|
7CWO
 
 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7FCC
 
 | | IL-1RAcPb TIR domain | | Descriptor: | Isoform 4 of Interleukin-1 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCJ
 
 | | Zebrafish SIGIRR TIR domain mutant - C299S | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
4JE0
 
 | |
3J1P
 
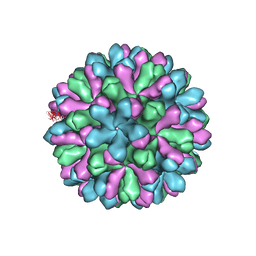 | | Atomic model of rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Liu, Y, Sun, F. | | Deposit date: | 2012-04-09 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
2P4H
 
 | |
4JDZ
 
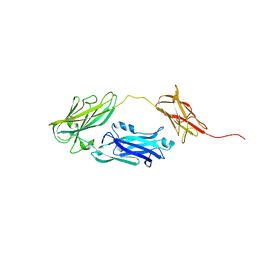 | |
6M0J
 
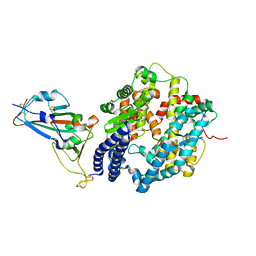 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
7F16
 
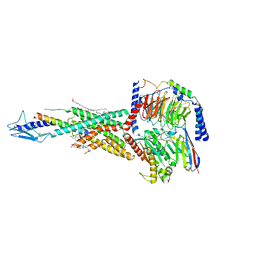 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
