8VOH
 
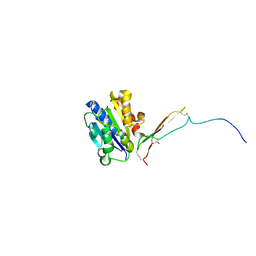 | |
7CWU
 
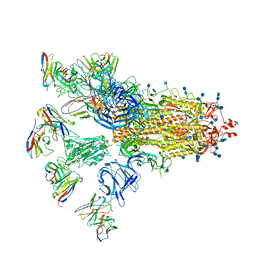 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7WT9
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab 9A8 | | Descriptor: | Heavy chain of Fab 9A8, Light chain of Fab 9A8, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
3J1P
 
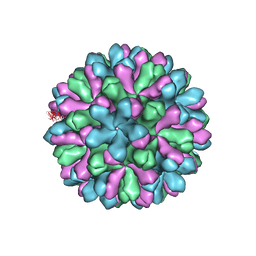 | | Atomic model of rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Liu, Y, Sun, F. | | Deposit date: | 2012-04-09 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
1LXF
 
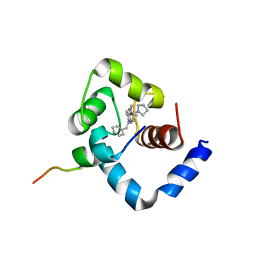 | | Structure of the Regulatory N-domain of Human Cardiac Troponin C in Complex with Human Cardiac Troponin-I(147-163) and Bepridil | | Descriptor: | 1-ISOBUTOXY-2-PYRROLIDINO-3[N-BENZYLANILINO] PROPANE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Sykes, B.D. | | Deposit date: | 2002-06-05 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the regulatory N-domain of human
cardiac troponin C in complex with human cardiac
troponin I147-163 and bepridil.
J.Biol.Chem., 277, 2002
|
|
3HBF
 
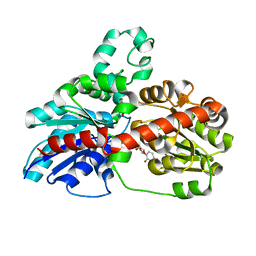 | | Structure of UGT78G1 complexed with myricetin and UDP | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
4Y35
 
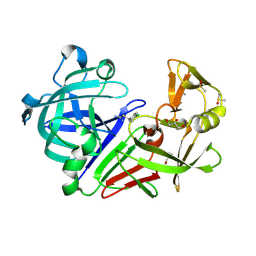 | | Endothiapepsin in complex with fragment 290 | | Descriptor: | 4-chlorobenzyl carbamimidothioate, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
7WEE
 
 | |
7WEB
 
 | |
7WE9
 
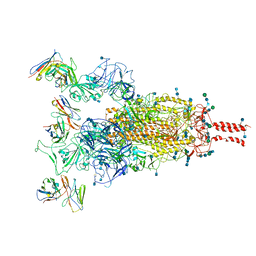 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
6M15
 
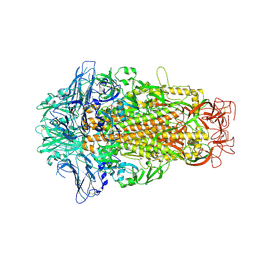 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
7WTK
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
2GYZ
 
 | | Crystal structure of human artemin | | Descriptor: | neurotrophic factor artemin isoform 3 | | Authors: | Wang, X.Q, Garcia, K.C. | | Deposit date: | 2006-05-10 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
4Y4A
 
 | | Endothiapepsin in complex with fragment 286 | | Descriptor: | 5-(aminomethyl)-2-methoxyphenol, ACETATE ION, Endothiapepsin, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y5E
 
 | | Endothiapepsin in complex with fragment 268 | | Descriptor: | (2R)-N-(trans-4-methylcyclohexyl)tetrahydrofuran-2-carboxamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.116 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
4Y58
 
 | |
4Y5K
 
 | | Endothiapepsin in complex with fragment 274 | | Descriptor: | (1S)-2-amino-1-(4-fluorophenyl)ethanol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
3VBR
 
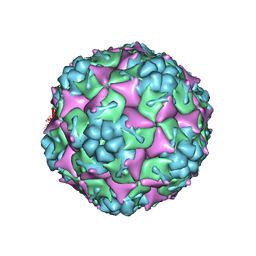 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6A7W
 
 | | Structure of a catalytic domain of the colistin resistance enzyme | | Descriptor: | Putative integral membrane protein, ZINC ION | | Authors: | Wang, X.D, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-07-04 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Structural and functional insights into MCR-2 mediated colistin resistance.
Sci China Life Sci, 61, 2018
|
|
4Y5G
 
 | |
8IDO
 
 | | Crystal structure of nanobody VHH-T148 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T148, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IDM
 
 | | Crystal structure of nanobody VHH-227 with nanobody VHH-T71 and MERS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8W90
 
 | | crystal structure of CD4-D1D2 with Nb457 | | Descriptor: | NB457, T-cell surface glycoprotein CD4 | | Authors: | Wang, X.Y, Wang, Y.X. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of CD4-D1D2 domain and nanobody Nb457 at 1.78 Angstroms resolution
To Be Published
|
|
3VBS
 
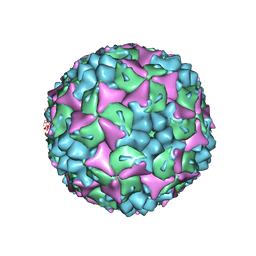 | | Crystal structure of human Enterovirus 71 | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
