7G9G
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with PCM-0102236-001 | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(2-methoxyphenyl)acetamide, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 実験手法 | X-RAY DIFFRACTION (2.079 Å) | | 主引用文献 | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G8Z
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1493056027 | | 分子名称: | 1-[(2R)-1-(methanesulfonyl)pyrrolidin-2-yl]methanamine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.509 Å) | | 主引用文献 | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G9H
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with PCM-0102245-001 | | 分子名称: | (2S)-4-(chloroacetyl)-3,4-dihydro-2H-1,4-benzoxazine-2-carboxamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 実験手法 | X-RAY DIFFRACTION (2.746 Å) | | 主引用文献 | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G82
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1079512010 | | 分子名称: | 6-methyl-2-[(3-methyl-1,2-oxazol-5-yl)methyl]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.409 Å) | | 主引用文献 | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G83
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z108545814 | | 分子名称: | 3-propanamido-1-benzofuran-2-carboxamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G8Y
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1449748885 | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(4-methoxyphenyl)glycinamide, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | 登録日 | 2023-06-22 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
5FTA
 
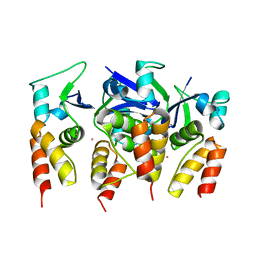 | | Crystal structure of the N-terminal BTB domain of human KCTD10 | | 分子名称: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 3, MERCURY (II) ION | | 著者 | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Tallant, C, Newman, J.A, Kopec, J, Fitzpatrick, F, Talon, R, Collins, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2016-01-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
6FT3
 
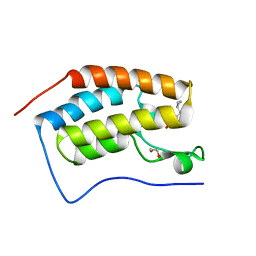 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FSY
 
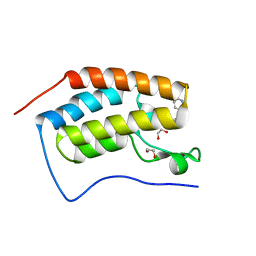 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
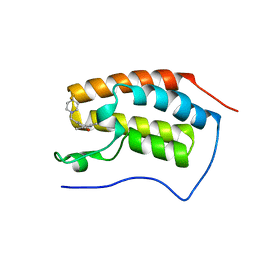 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6G0R
 
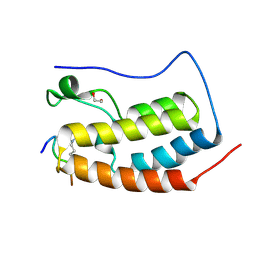 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated POLR2A peptide (K775ac/K778ac) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DNA-directed RNA polymerase II subunit RPB1 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-03-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5FPV
 
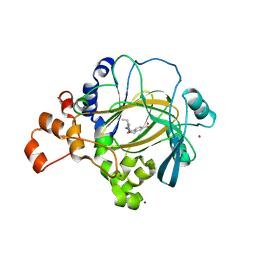 | | Crystal structure of human JMJD2A in complex with compound KDOAM20A | | 分子名称: | 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, MANGANESE (II) ION, ... | | 著者 | Srikannathasan, V, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | 登録日 | 2015-12-03 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
7FQZ
 
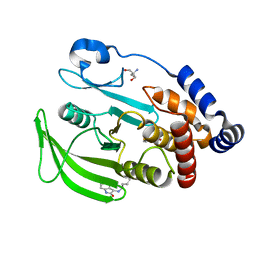 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000203a | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-fluoro-1,3-dihydro-2H-indol-2-one, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | 登録日 | 2022-10-20 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
5S3Q
 
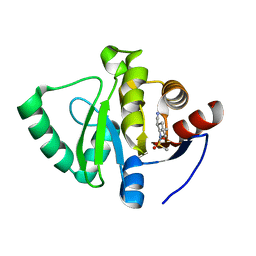 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0013 | | 分子名称: | (2R,3R)-2-methyl-1-(methylsulfonyl)piperidine-3-carbonitrile, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S1O
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with STL414928 | | 分子名称: | 2H-pyrazolo[3,4-b]pyridin-5-amine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S42
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00023833 | | 分子名称: | 4-bromanyl-1,8-naphthyridine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S8O
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N01460c (space group P212121) | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, N-(4-chloro-2-methylphenyl)acetamide, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | 登録日 | 2020-12-17 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | XChem group deposition
To Be Published
|
|
5S6X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2889976755 | | 分子名称: | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S1A
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-43406 | | 分子名称: | 5-amino-3-methyl-1H-pyrazole-4-carbonitrile, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.079 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3F
 
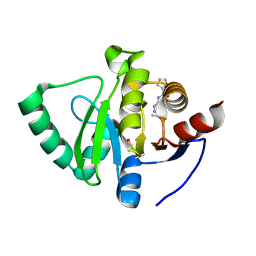 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z57446103 | | 分子名称: | DIMETHYL SULFOXIDE, N-(2-propyl-2H-tetrazol-5-yl)furan-2-carboxamide, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4B
 
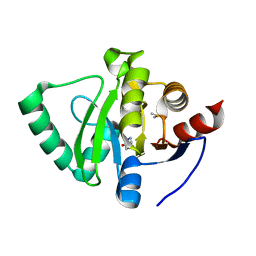 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z3219959731 | | 分子名称: | DIMETHYL SULFOXIDE, Non-structural protein 3, pyridazin-3(2H)-one | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.185 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S70
 
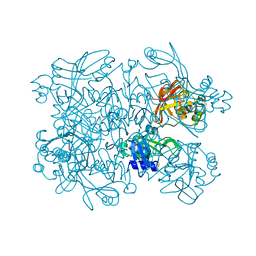 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | 分子名称: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.327 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S2F
 
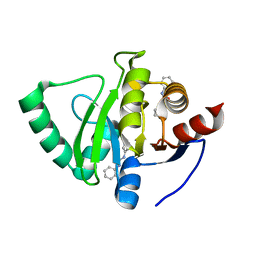 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z44592329 | | 分子名称: | N-phenyl-N'-pyridin-3-ylurea, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.186 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3A
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1562205518 | | 分子名称: | 1-(2-hydroxyethyl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.178 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4E
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z2301685688 | | 分子名称: | 1H-imidazole-5-carbonitrile, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
