1LPA
 
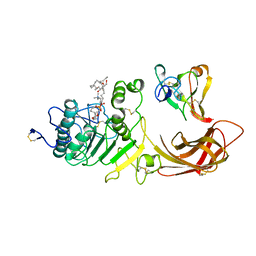 | | INTERFACIAL ACTIVATION OF THE LIPASE-PROCOLIPASE COMPLEX BY MIXED MICELLES REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, COLIPASE, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Van Tilbeurgh, H, Egloff, M.-P, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography.
Nature, 362, 1993
|
|
1B56
 
 | | HUMAN RECOMBINANT EPIDERMAL FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Van Tilbeurgh, H, Hohoff, C, Borchers, T, Spener, F. | | Deposit date: | 1999-01-12 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein.
Biochemistry, 38, 1999
|
|
1N8S
 
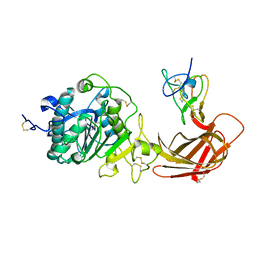 | | Structure of the pancreatic lipase-colipase complex | | Descriptor: | Triacylglycerol lipase, pancreatic, colipase II | | Authors: | van Tilbeurgh, H, Sarda, L, Verger, R, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the pancreatic lipase-procolipase complex
Nature, 359, 1992
|
|
1H99
 
 | |
1H54
 
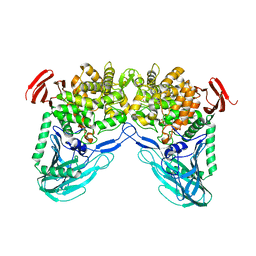 | | Maltose phosphorylase from Lactobacillus brevis | | Descriptor: | MALTOSE PHOSPHORYLASE, PHOSPHATE ION, POTASSIUM ION | | Authors: | Van Tilbeurgh, H, Egloff, M.-P. | | Deposit date: | 2001-05-18 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Maltose Phosphorylase from Lactobacillus Brevis: Unexpected Evolutionary Relationship with Glucoamylases.
Structure, 9, 2001
|
|
6Z81
 
 | | TsaBD bound to the inhibitor | | Descriptor: | (2~{S},3~{R})-2-[2-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]ethanoylamino]-3-oxidanyl-butanoic acid, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Missoury, S, Van Tilbeurgh, H. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a reaction intermediate mimic in t6A biosynthesis bound in the active site of the TsaBD heterodimer from Escherichia coli.
Nucleic Acids Res., 49, 2021
|
|
1QVV
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVW
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
4ML3
 
 | | X-ray structure of ComE D58A REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
4MLD
 
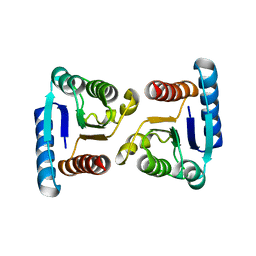 | | X-ray structure of ComE D58E REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
1LPB
 
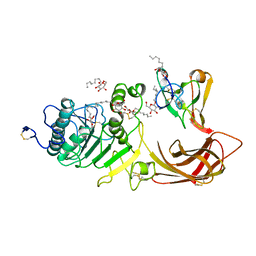 | | THE 2.46 ANGSTROMS RESOLUTION STRUCTURE OF THE PANCREATIC LIPASE COLIPASE COMPLEX INHIBITED BY A C11 ALKYL PHOSPHONATE | | Descriptor: | CALCIUM ION, COLIPASE, LIPASE, ... | | Authors: | Egloff, M.-P, Van Tilbeurgh, H, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Biochemistry, 34, 1995
|
|
5CM2
 
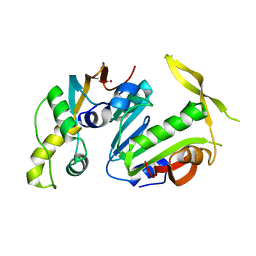 | | Structure of Y. lipolytica Trm9-Trm112 complex, a methyltransferase modifying U34 in the anticodon loop of some tRNAs | | Descriptor: | TRNA METHYLTRANSFERASE, TRNA METHYLTRANSFERASE ACTIVATOR SUBUNIT, ZINC ION | | Authors: | Letoquart, J, van Tran, N, Caroline, V, Aleksandrov, A, Lazar, N, Van Tilbeurgh, H, Liger, D, Graille, M. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure.
Nucleic Acids Res., 43, 2015
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7AD5
 
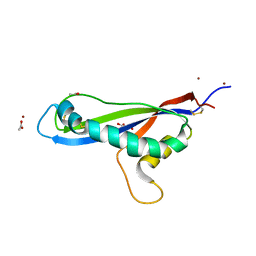 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7B76
 
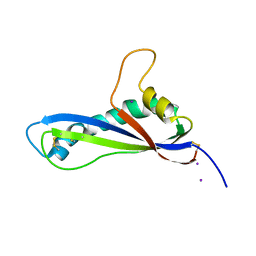 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | Avirulence protein LmJ1, IODIDE ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
5DCQ
 
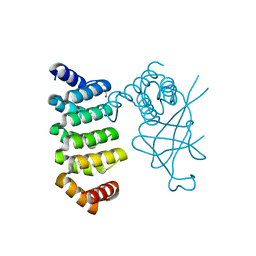 | | Crystal structure of bacterial adhesin, FNE from Streptococcus equi spp. equi. | | Descriptor: | FORMIC ACID, Fibronectin-binding protein, artificial repeat proteins (alphaREP3) | | Authors: | Tiouajni, M, Graille, M, van Tilbeurgh, H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and functional analysis of the fibronectin-binding protein FNE from Streptococcus equi spp. equi.
FEBS J., 281, 2014
|
|
6XUI
 
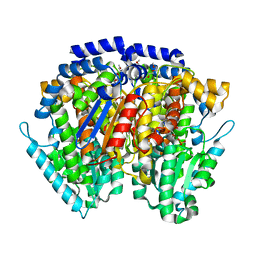 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5-PHOSPHOARABINONIC ACID, GLYCEROL, ... | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2020-01-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
6XUH
 
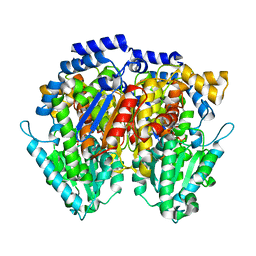 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | Descriptor: | (2R,3R,4S)-5-((2-aminoethyl)amino)-2,3,4-trihydroxy-5-oxopentyl dihydrogen phosphate, 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2020-01-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
4WW9
 
 | |
5MTZ
 
 | | Crystal structure of a long form RNase Z from yeast | | Descriptor: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | Authors: | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-06-21 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
4WWA
 
 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WW7
 
 | |
4WX8
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1 | | Authors: | Zhang, W, Van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WXA
 
 | | Crystal structure of binary complex Gon7-Pcc1 | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit GON7, EKC/KEOPS complex subunit PCC1, ... | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
