5N83
 
 | |
5N8D
 
 | |
5M9F
 
 | |
2BSF
 
 | | Structure of the C-terminal receptor-binding domain of avian reovirus fibre sigmaC, Zn crystal form. | | Descriptor: | SIGMA C CAPSID PROTEIN, SULFATE ION, ZINC ION | | Authors: | Guardado Calvo, P, Fox, G.C, Hermo Parrado, X.L, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Carboxy-Terminal Receptor-Binding Domain of Avian Reovirus Fibre Sigmac
J.Mol.Biol., 354, 2005
|
|
4CT3
 
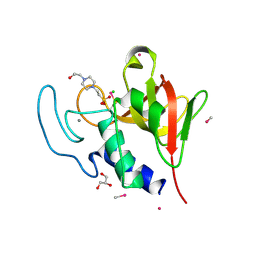 | | Methylmercury chloride derivative structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
4A0U
 
 | |
4CSH
 
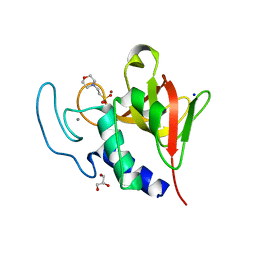 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
4D1G
 
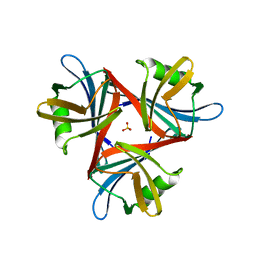 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, second P212121 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4A0T
 
 | |
4D1F
 
 | |
4D0U
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, selenomethionine-derivative | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
4D0V
 
 | | Crystal structure of the fiber head domain of the Atadenovirus snake adenovirus 1, native, I213 crystal form | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FIBER PROTEIN, SULFATE ION | | Authors: | Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the fibre head domain of the Atadenovirus Snake Adenovirus 1.
PLoS ONE, 9, 2014
|
|
1PDL
 
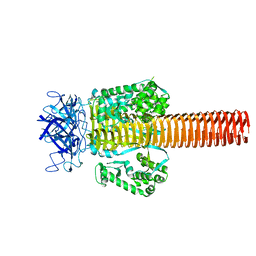 | | Fitting of gp5 in the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Tail-associated lysozyme | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDJ
 
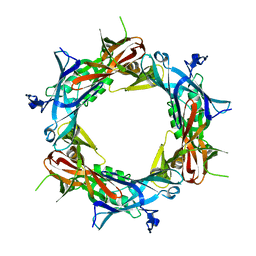 | | Fitting of gp27 into cryoEM reconstruction of bacteriophage T4 baseplate | | Descriptor: | Baseplate structural protein Gp27 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDP
 
 | | Fitting of gp9 structure into the bacteriophage T4 baseplate cryoEM reconstruction | | Descriptor: | Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
2JJL
 
 | | Structure of avian reovirus sigmaC117-326, P321 crystal form | | Descriptor: | CHLORIDE ION, SIGMA-C CAPSID PROTEIN, SULFATE ION, ... | | Authors: | Guardado-Calvo, P, Fox, G.C, Llamas-Saiz, A.L, Benavente, J, van Raaij, M.J. | | Deposit date: | 2008-04-14 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre.
J. Gen. Virol., 90, 2009
|
|
1PDF
 
 | | Fitting of gp11 crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 baseplate-tail tube complex | | Descriptor: | Baseplate structural protein Gp11 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDM
 
 | | Fitting of gp8 structure into the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Baseplate structural protein Gp8 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
2WYU
 
 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase apo-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, GLYCEROL, SODIUM ION | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
2WYV
 
 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase NAD-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
2WYW
 
 | | High resolution structure of Thermus thermophilus enoyl-acyl carrier protein reductase NAD and triclosan-form | | Descriptor: | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Otero, J.M, Noel, A.J, Guardado-Calvo, P, Llamas-Saiz, A.L, van Raaij, M.J. | | Deposit date: | 2009-11-20 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of Thermus thermophilus enoyl-acyl carrier protein reductase in the apo form, in complex with NAD+ and in complex with NAD+ and triclosan.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
2Y77
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-2-ylmethoxy)-1,4,5- trihydroxy-2-(thiophen-2-ylmethyl)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[B]THIOPHEN-2-YL)METHOXY-1,4,5-TRIHYDROXY-2-(THIEN-2-YL)METHYLCYCLOHEX-2-EN-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2Y71
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-1,4,5-trihydroxy-3-((5-methylbenzo(b) thiophen-2-yl)methoxy)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2Y76
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-5-ylmethoxy)-2-(benzo(b) thiophen-5-ylmethyl)-1,4,5-trihydroxycyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[b]THIOPHEN-5-YL)METHOXY-2-(BENZO[b]THIOPHEN-5-YL)METHYL-1,4,5-TRIHYDROXYCYCLOHEX-2-ENE-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2XDA
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-2-(2-Cyclopropyl)ethyl-4,6,7- trihydroxy-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-2-(2-CYCLOPROPYLETHYL)-4,6,7-TRIHYDROXY-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
