7DZK
 
 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
6JBZ
 
 | |
8WYD
 
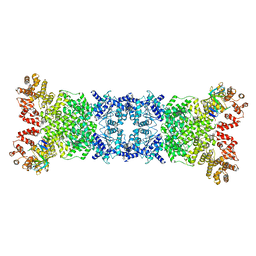 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY9
 
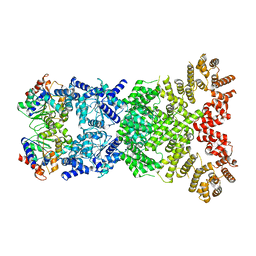 | |
8WYE
 
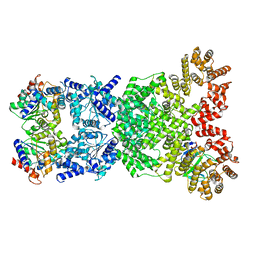 | | Cryo-EM structure of DSR2-DSAD1 (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
3S2S
 
 | |
3S70
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO solved by As-SAD | | Descriptor: | ACETATE ION, CACODYLATE ION, Caspase-6, ... | | Authors: | Su, X.-D, Liu, X, Wang, X.-J. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer
Plos One, 6, 2011
|
|
8WYA
 
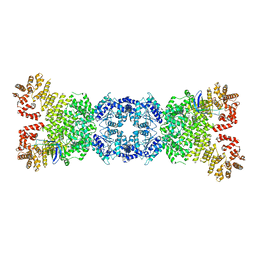 | | Cryo-EM structure of DSR2-tube complex | | Descriptor: | Bacillus phage SPbeta tube protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYB
 
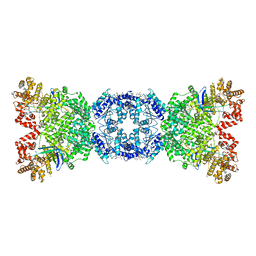 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ complex | | Descriptor: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY8
 
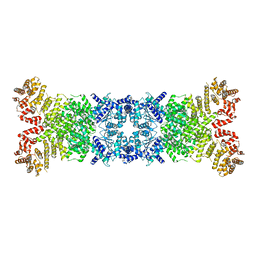 | | Cryo-EM structure of DSR2 apo complex | | Descriptor: | SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
6A9W
 
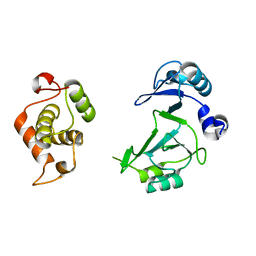 | | Structure of the bifunctional DNA primase-polymerase from phage NrS-1 | | Descriptor: | Primase | | Authors: | Guo, H.J, Li, M.J, Wang, T.L, Wu, H, Zhou, H, Xu, C.Y, Liu, X.P, Yu, F, He, J.H. | | Deposit date: | 2018-07-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and biochemical studies of the bifunctional DNA primase-polymerase from phage NrS-1.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
8WYF
 
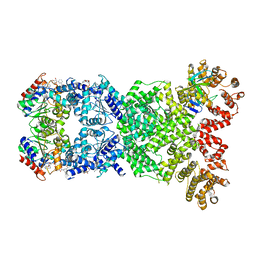 | | Cryo-EM structure of DSR2-DSAD1-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYC
 
 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
7F05
 
 | |
4MH1
 
 | |
9EQM
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-21 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[trans-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
6IG5
 
 | |
6IGA
 
 | |
6IV7
 
 | | The crystal structure of a SAM-dependent enzyme from aspergillus flavus | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase lepI | | Authors: | Zhou, Y.Z, Peng, T, Liao, L.J, Liu, X.K, Zhao, Y.C, Guo, Y, Zeng, Z.X. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | The crystal structure of A SAM-Dependent enzyme from Aspergillus flavus
To Be Published
|
|
6IY3
 
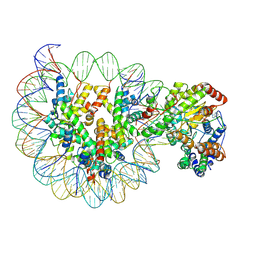 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6IY2
 
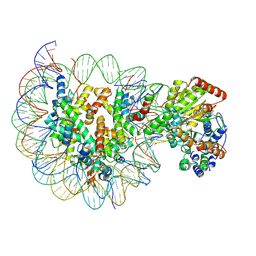 | | Structure of Snf2-MMTV-A nucleosome complex at shl2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
2JOO
 
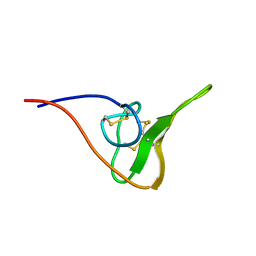 | | The NMR Solution Structure of Recombinant RGD-hirudin | | Descriptor: | Hirudin variant-1 | | Authors: | Song, X, Mo, W, Liu, X, Yan, X, Song, H, Dai, L. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of recombinant RGD-hirudin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
6A9Y
 
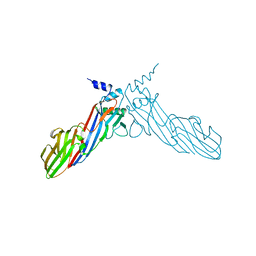 | | The crystal structure of Mu homology domain of SGIP1 | | Descriptor: | SH3-containing GRB2-like protein 3-interacting protein 1 | | Authors: | Feng, Y, Liu, X. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SGIP1 dimerizes via intermolecular disulfide bond in mu HD domain during cellular endocytosis.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
