6Y9A
 
 | |
6FC5
 
 | | Bik1 CAP-Gly domain | | Descriptor: | Microtubule-associated protein | | Authors: | Kumar, A, Stangier, M.M, Steinmetz, M.O. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Function Relationship of the Bik1-Bim1 Complex.
Structure, 26, 2018
|
|
6Y97
 
 | |
6FC6
 
 | |
3ND9
 
 | |
8JCS
 
 | |
8JCQ
 
 | |
8JCR
 
 | | Crystal structure of Procerain from Calotropis gigantea (pH 6.0) | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Procerain from Calotropis gigantea
To Be Published
|
|
8OH9
 
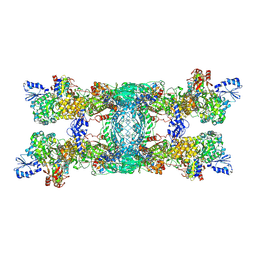 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
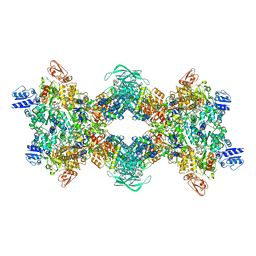 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
7RBN
 
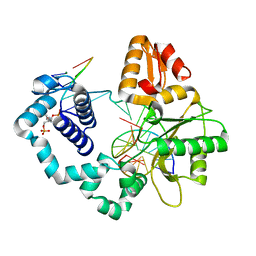 | | Human DNA polymerase beta crosslinked complex, 20 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBJ
 
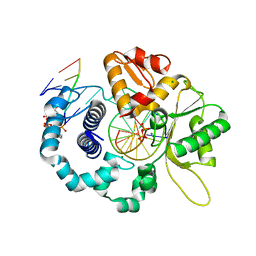 | | Human DNA polymerase beta crosslinked complex, 30 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBI
 
 | | Human DNA polymerase beta crosslinked complex, 20 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBO
 
 | | Human DNA polymerase beta crosslinked complex, 60 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBH
 
 | | Human DNA polymerase beta crosslinked ternary complex 2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBL
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBM
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mn exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBK
 
 | | Human DNA polymerase beta crosslinked complex, 40 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5ID2
 
 | |
7F8R
 
 | |
5EVO
 
 | | Structure of Dehydroascrobate Reductase from Pennisetum Americanum in complex with two non-native ligands, Acetate in the G-site and Glycerol in the H-site | | Descriptor: | ACETATE ION, Dehydroascorbate reductase, GLYCEROL | | Authors: | Kumar, A.O, Das, B.K, Arockiasamy, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
8BR4
 
 | |
7VMF
 
 | |
4DJJ
 
 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution | | Descriptor: | PIMELIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution
To be Published
|
|
1JYM
 
 | | Crystals of Peptide Deformylase from Plasmodium falciparum with Ten Subunits per Asymmetric Unit Reveal Critical Characteristics of the Active Site for Drug Design | | Descriptor: | COBALT (II) ION, Peptide Deformylase | | Authors: | Kumar, A, Nguyen, K.T, Srivathsan, S, Ornstein, B, Turley, S, Hirsh, I, Pei, D, Hol, W.G.J. | | Deposit date: | 2001-09-12 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design.
Structure, 10, 2002
|
|
