4JX9
 
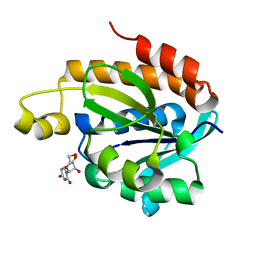 | | Crystal structure of the complex of peptidyl t-RNA hydrolase from Acinetobacter baumannii with uridine at 1.4A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, URIDINE | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-28 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
3MRY
 
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution
To be Published
|
|
4JTB
 
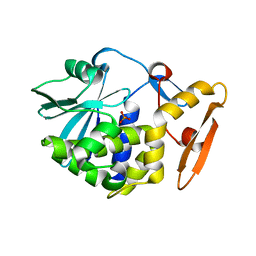 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, rRNA N-glycosidase | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-23 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution
To be published
|
|
3MRW
 
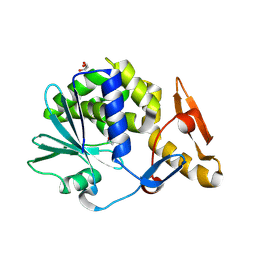 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
4JY7
 
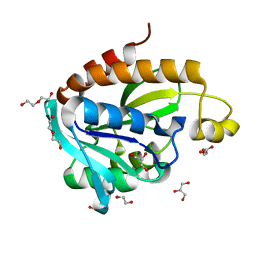 | | Crystal structure of Acinetobacter baumannii Peptidyl-tRNA Hydrolase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamini, S, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
3N31
 
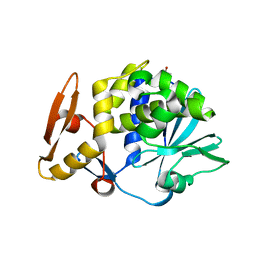 | | Crystal Structure of the complex of type I ribosome inactivating protein with fucose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Perbandt, M, Betzel, C, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fucose at 2.1A resolution
To be Published
|
|
3MY6
 
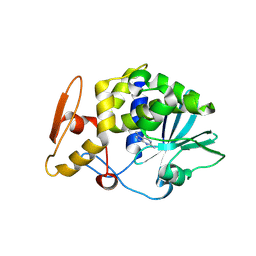 | | Crystal Structure of the complex of type 1 ribosome inactivating protein with 7-methylguanine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-7-methyl-1,7-dihydro-6H-purin-6-one, Ribosome-inactivating protein momordin I | | Authors: | Kushwaha, G.S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-10 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the complex of type 1 ribosome inactivating protein with 7-methylguanine at 2.65 A resolution
To be Published
|
|
3N1D
 
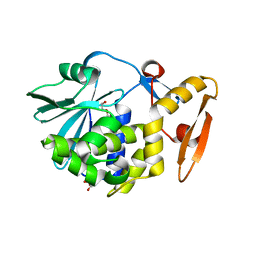 | | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution
To be Published
|
|
3N2D
 
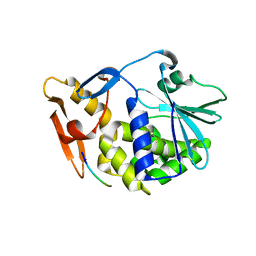 | | Crystal Structure of the Complex of type I Ribosome inactivating protein with hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-meric peptide from 60S acidic ribosomal protein P2-beta, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Prem Kumar, R, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of the Complex of type I Ribosome inactivating protein with hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 2.2 A resolution
To be Published
|
|
3CXA
 
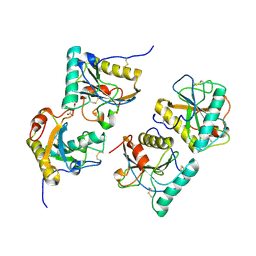 | | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Balaji, K, Sharma, P, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution
To be Published
|
|
3N5D
 
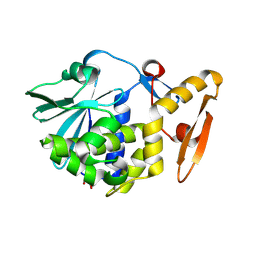 | | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution
To be Published
|
|
3FAQ
 
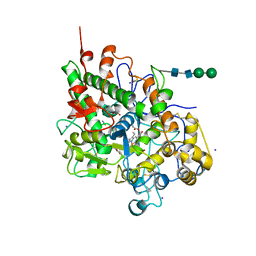 | | Crystal structure of lactoperoxidase complex with cyanide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3N1N
 
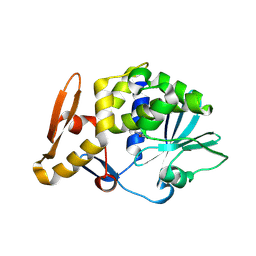 | | Crystal structure of the complex of type I ribosome inactivating protein with guanine at 2.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Singh, N, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-16 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
3FNL
 
 | | Crystal Structure of the Complex of Buffalo Lactoperoxidase with Salicylhydroxamic Acid at 2.48 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Sheikh, I.A, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-12-25 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of the Complex of Buffalo Lactoperoxidase with Salicylhydroxamic Acid at 2.48 A Resolution
To be Published
|
|
3N3X
 
 | | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Vikram, G, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution
To be Published
|
|
3NJS
 
 | | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution
To be Published
|
|
3NFM
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution
To be Published
|
|
5Z1M
 
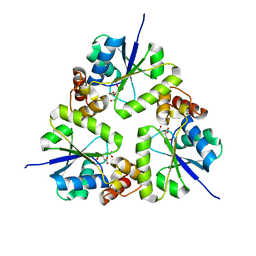 | | Crystal structure of the complex of trimeric Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate ion at 1.87 A resolution | | Descriptor: | CITRIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-12-26 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the complex of trimeric Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate ion at 1.87 A resolution
To Be Published
|
|
3NX9
 
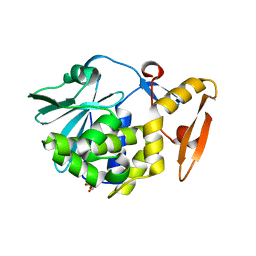 | | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-13 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution
To be Published
|
|
4IKO
 
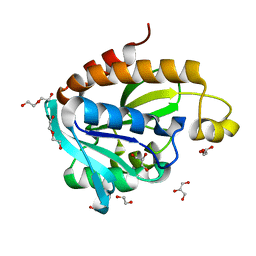 | | Structure of Peptidyl- tRNA Hydrolase from Acinetobacter baumannii at 1.90 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamini, S, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-12-27 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
3COR
 
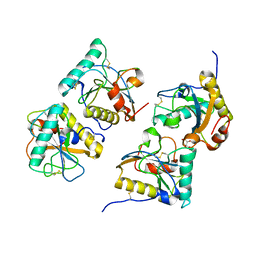 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with N-acetylgalactosamine at 3.1 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein | | Authors: | Sharma, P, Vikram, G, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with N-acetylgalactosamine at 3.1 A resolution
To be Published
|
|
4JWK
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Acinetobacter baumannii with cytidine at 1.87 A resolution | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
5ZZC
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 1.94A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Gupta, A, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-05-31 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 1.94 A resolution
To Be Published
|
|
3GCL
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid
J.Biol.Chem., 284, 2009
|
|
4LJ2
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 3.15A resolution | | Descriptor: | Chorismate synthase | | Authors: | Chaudhary, A, Singh, N, Kaushik, S, Tyagi, T.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-04 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 3.15A resolution
To be Published
|
|
