4OB9
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Shukla, P.K, Chaudhary, A, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To be Published
|
|
4G2Z
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Mefenamic acid at 1.90 A Resolution | | Descriptor: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Mefenamic acid at 1.90 A Resolution
To be Published
|
|
3MJN
 
 | | Crystal Structure of the complex of C-lobe of lactoferrin with isopropylamino-3-(1-naphthyloxy)propan-2-ol at 2.38 A Resolution | | Descriptor: | (1E,2R)-1-(ISOPROPYLIMINO)-3-(1-NAPHTHYLOXY)PROPAN-2-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Srivastava, K, Vikram, G, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the complex of C-lobe of lactoferrin with isopropylamino-3-(1-naphthyloxy)propan-2-ol at 2.38 A Resolution
To be Published
|
|
4GM7
 
 | | Structure of cinnamic acid bound bovine lactoperoxidase at 2.6A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cinnamic acid bound bovine lactoperoxidase at 2.6A resolution
To be Published
|
|
3M7S
 
 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
3MRW
 
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
4GLB
 
 | | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution | | Descriptor: | 4-nitrobenzaldehyde, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution
TO BE PUBLISHED
|
|
4GLD
 
 | | Crystal Structure of the complex of type II phospholipase A2 with a designed peptide inhibitor Phe - Leu - Ala - Tyr - Lys at 1.69 A resolution | | Descriptor: | ACETATE ION, FLAYK, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Shukla, P.K, Sinha, M, Dey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of the complex of type II phospholipase A2 with a designed peptide inhibitor Phe - Leu - Ala - Tyr - Lys at 1.69 A resolution
To be Published
|
|
3NX9
 
 | | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-13 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of type I ribosome inactivating protein in complex with maltose at 1.7A resolution
To be Published
|
|
4HMB
 
 | | Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Haridas, M, Chandra, D.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution
To be Published
|
|
3OSZ
 
 | | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution | | Descriptor: | 10-mer peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-10 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution
To be Published
|
|
3OIH
 
 | | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, M, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution
To be Published
|
|
3MRY
 
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution
To be Published
|
|
4MSF
 
 | | Crystal structure of the complex of goat lactoperoxidase with 3-hydroxymethyl phenol at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(hydroxymethyl)phenol, ... | | Authors: | Singh, A, Singh, R.P, Sinha, M, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with 3-hydroxymethyl phenol at 1.98 Angstrom resolution
To be published
|
|
4GF9
 
 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
4GFY
 
 | | Design of peptide inhibitors of phospholipase A2: crystal Structure of phospholipase A2 complexed with a designed tetrapeptide Val - Ilu- Ala - Lys at 2.7 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, VIAK | | Authors: | Shukla, P.K, Sinha, M, Dey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-04 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of peptide inhibitors of phospholipase A2: crystal Structure of phospholipase A2 complexed with a designed tetrapeptide Val - Ilu- Ala - Lys at 2.7 A resolution
To be Published
|
|
4JC4
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2013-02-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4GRK
 
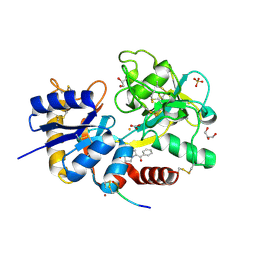 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with ketorolac at 1.68 A Resolution | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gautam, L, Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-25 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with ketorolac at 1.68 A Resolution
To be Published
|
|
4GWL
 
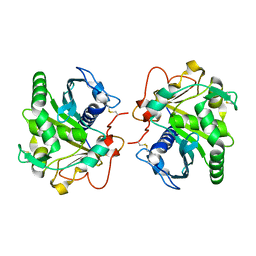 | | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.55A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Shukla, P.K, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-09-03 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.55A resolution
To be Published
|
|
3N5D
 
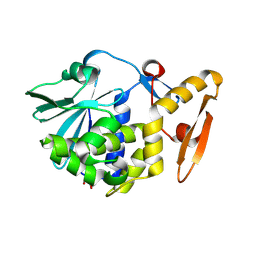 | | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution
To be Published
|
|
3N1N
 
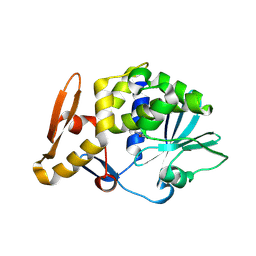 | | Crystal structure of the complex of type I ribosome inactivating protein with guanine at 2.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Singh, N, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-16 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
3NJS
 
 | | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution
To be Published
|
|
3N3X
 
 | | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Vikram, G, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution
To be Published
|
|
3NFM
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution
To be Published
|
|
3N31
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fucose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Perbandt, M, Betzel, C, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fucose at 2.1A resolution
To be Published
|
|
