6UTI
 
 | |
7DY7
 
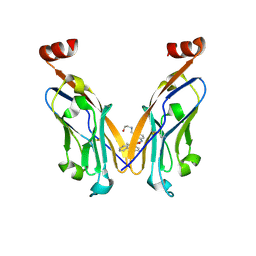 | | Discovery of Novel Small-molecule Inhibitors of PD-1/PD-L1 Axis that Promotes PD-L1 Internalization and Degradation | | 分子名称: | 2-[[3-[[5-(2-methyl-3-phenyl-phenyl)-1,3,4-oxadiazol-2-yl]amino]phenyl]methylamino]ethanol, Programmed cell death 1 ligand 1 | | 著者 | Cheng, Y, Wang, T.Y, Lu, M.L, Jiang, S, Xiao, Y.B. | | 登録日 | 2021-01-20 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation.
J.Med.Chem., 65, 2022
|
|
6UTF
 
 | |
5GKB
 
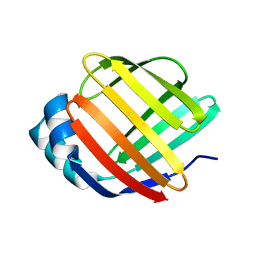 | | Crystal Structure of Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster without citrate inside | | 分子名称: | Fatty acid bindin protein, isoform B | | 著者 | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | 登録日 | 2016-07-04 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
5GGE
 
 | | Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster | | 分子名称: | CITRIC ACID, Fatty acid bindin protein, isoform B | | 著者 | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | 登録日 | 2016-06-15 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.861 Å) | | 主引用文献 | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
4XRB
 
 | |
1M08
 
 | | Crystal structure of the unbound nuclease domain of ColE7 | | 分子名称: | Colicin E7, PHOSPHATE ION, ZINC ION | | 著者 | Cheng, Y.S, Hsia, K.C, Doudeva, L.G, Chak, K.F, Yuan, H.S. | | 登録日 | 2002-06-12 | | 公開日 | 2002-12-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of the Nuclease Domain of Colicin E7 Suggests a Mechanism for Binding to Double-stranded DNA by the H-N-H Endonucleases
J.mol.biol., 324, 2002
|
|
3VHO
 
 | | Y61-gg insertion mutant of Tm-Cellulase 12A | | 分子名称: | Endo-1,4-beta-glucanase | | 著者 | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | 登録日 | 2011-08-30 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3VHN
 
 | | Y61G mutant of Cellulase 12A from thermotoga maritima | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | 登録日 | 2011-08-30 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3VHP
 
 | | The insertion mutant Y61GG of Tm Cel12A | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | 登録日 | 2011-08-30 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | 分子名称: | Endo-1,4-beta-glucanase | | 著者 | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | 登録日 | 2010-08-20 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMP
 
 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | 登録日 | 2010-08-20 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
1W0H
 
 | | Crystallographic structure of the nuclease domain of 3'hExo, a DEDDh family member, bound to rAMP | | 分子名称: | 3'-5' EXONUCLEASE ERI1, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION | | 著者 | Cheng, Y, Patel, D. | | 登録日 | 2004-06-04 | | 公開日 | 2004-09-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystallographic Structure of the Nuclease Domain of 3'Hexo, a Deddh Family Member, Bound to Ramp
J.Mol.Biol., 343, 2004
|
|
1UOV
 
 | | Calcium binding domain C2B | | 分子名称: | CALCIUM ION, GLYCEROL, SYNAPTOTAGMIN I | | 著者 | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | 登録日 | 2003-09-24 | | 公開日 | 2004-09-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1UOW
 
 | | Calcium binding domain C2B | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | 登録日 | 2003-09-24 | | 公開日 | 2004-09-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | 登録日 | 2010-08-20 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | 登録日 | 2010-08-20 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
1TJM
 
 | | Crystallographic Identification of Sr2+ Coordination Site in Synaptotagmin I C2B Domain | | 分子名称: | GLYCEROL, STRONTIUM ION, Synaptotagmin I | | 著者 | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | 登録日 | 2004-06-06 | | 公開日 | 2004-09-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain
Protein Sci., 13, 2004
|
|
3AMQ
 
 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | 登録日 | 2010-08-20 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
1ETX
 
 | |
1ETK
 
 | |
1ETY
 
 | |
1ETO
 
 | |
1ETQ
 
 | |
1JE9
 
 | | NMR SOLUTION STRUCTURE OF NT2 | | 分子名称: | SHORT NEUROTOXIN II | | 著者 | Cheng, Y, Wang, W, Wang, J. | | 登録日 | 2001-06-16 | | 公開日 | 2001-07-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
