1W9B
 
 | | S. alba myrosinase in complex with carba-glucotropaeolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBA-GLUCOTROPAEOLIN, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The glucosinolate-myrosinase system. New insights into enzyme-substrate interactions by use of simplified inhibitors.
Org. Biomol. Chem., 3, 2005
|
|
1W9D
 
 | | S. alba myrosinase in complex with S-ethyl phenylacetothiohydroximate- O-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Glucosinolate-Myrosinase System. New Insights Into Enzyme-Substrate Interactions by Use of Simplified Inhibitors
Org.Biomol.Chem., 3, 2005
|
|
2WXD
 
 | | A MICROMOLAR O-SULFATED THIOHYDROXIMATE INHIBITOR BOUND TO PLANT MYROSINASE | | Descriptor: | 2-(DIMETHYLAMINO)ETHYL (1Z)-2-PHENYL-N-(SULFOOXY)ETHANIMIDOTHIOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Besle, A, Burmeister, W.P. | | Deposit date: | 2009-11-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Micromolar O-Sulfated Thiohydroximate Inhibitor Bound to Plant Myrosinase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1WCG
 
 | | Aphid myrosinase | | Descriptor: | GLYCEROL, THIOGLUCOSIDASE | | Authors: | Husebye, H, Arzt, S, Burmeister, W.P, Haertel, F.V, Brandt, A, Rossiter, J.T, Bones, A.M. | | Deposit date: | 2004-11-15 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure at 1.1A Resolution of an Insect Myrosinase from Brevicoryne Brassicae Shows its Close Relationship to Beta-Glucosidases.
Insect Biochem.Mol.Biol., 35, 2005
|
|
1O6E
 
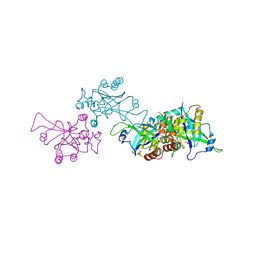 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
3FRU
 
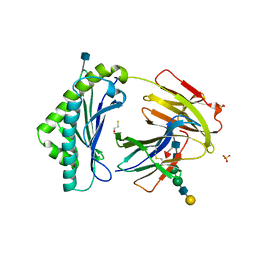 | | NEONATAL FC RECEPTOR, PH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, BETA-MERCAPTOETHANOL, ... | | Authors: | Vaughn, D.E, Burmeister, W.P, Bjorkman, P.J. | | Deposit date: | 1997-12-22 | | Release date: | 1998-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pH-dependent antibody binding by the neonatal Fc receptor.
Structure, 6, 1998
|
|
4OD8
 
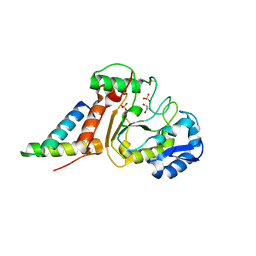 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
4ODA
 
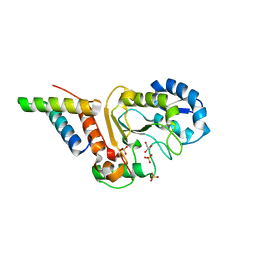 | | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminus | | Descriptor: | DNA polymerase processivity factor component A20, GLYCEROL, SULFATE ION, ... | | Authors: | Contesto-Richefeu, C, Tarbouriech, N, Brazzolotto, X, Burmeister, W.P, Iseni, F. | | Deposit date: | 2014-01-10 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Plos Pathog., 10, 2014
|
|
1EZJ
 
 | | CRYSTAL STRUCTURE OF THE MULTIMERIZATION DOMAIN OF THE PHOSPHOPROTEIN FROM SENDAI VIRUS | | Descriptor: | CALCIUM ION, ETHYL MERCURY ION, NUCLEOCAPSID PHOSPHOPROTEIN | | Authors: | Tarbouriech, N, Curran, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2000-05-11 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric coiled coil domain of Sendai virus phosphoprotein.
Nat.Struct.Biol., 7, 2000
|
|
