6QST
 
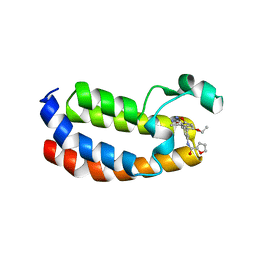 | | Structure of CREBBP bromodomain with compound 2 bound | | Descriptor: | CREB-binding protein, ~{N}-[3-(3-azanyl-5-methyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Sledz, P, Caflisch, A. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of CREBBP bromodomain with compound 2 bound
To Be Published
|
|
2KBK
 
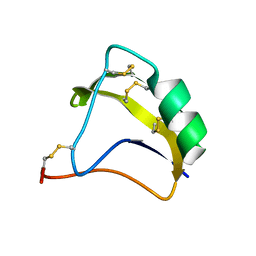 | | Solution Structure of BmK-M10 | | Descriptor: | Neurotoxin BmK-M10 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmK-M10
To be Published
|
|
2KBJ
 
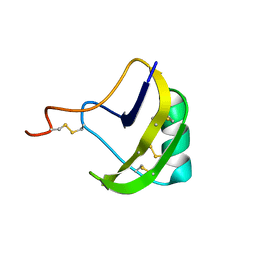 | | solution structure of BmKalphaTx11 (minor conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2KBH
 
 | | solution structure of BmKalphaTx11 (major conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2P3E
 
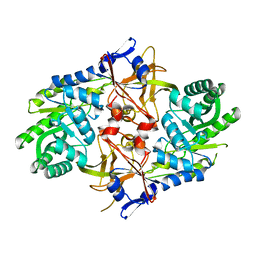 | | Crystal structure of AQ1208 from Aquifex aeolicus | | Descriptor: | Diaminopimelate decarboxylase | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be Published
To be Published
|
|
2P17
 
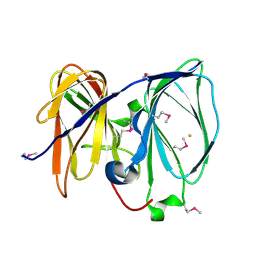 | | Crystal structure of GK1651 from Geobacillus kaustophilus | | Descriptor: | FE (III) ION, Pirin-like protein | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-05-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of GK1651 from Geobacillus kaustophilus
To be Published
|
|
4V7P
 
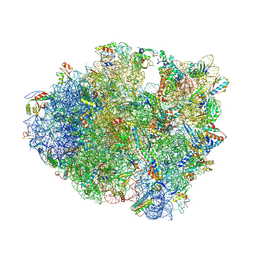 | | Recognition of the amber stop codon by release factor RF1. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Zhu, J, Asahara, H, Noller, H.F. | | Deposit date: | 2010-04-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition of the amber UAG stop codon by release factor RF1.
Embo J., 29, 2010
|
|
2FQO
 
 | | Crystal structure of B. subtilis LuxS in complex with (2S)-2-Amino-4-[(2R,3R)-2,3-dihydroxy-3-N- hydroxycarbamoyl-propylmercapto]butyric acid | | Descriptor: | (2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYCARBAMOYL-PROPYLMERCAPTO]BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteine lyase, ... | | Authors: | Shen, G, Rajan, R, Zhu, J, Bell, C.E, Pei, D. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design and Synthesis of Substrate and Intermediate Analogue Inhibitors of S-Ribosylhomocysteinase
J.Med.Chem., 49, 2006
|
|
2FQT
 
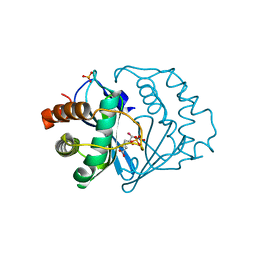 | | Crystal structure of B.subtilis LuxS in complex with (2S)-2-Amino-4-[(2R,3S)-2,3-dihydroxy-3-N-hydroxycarbamoyl-propylmercapto]butyric acid | | Descriptor: | (2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYCARBAMOYL-PROPYLMERCAPTO]BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteine lyase, ... | | Authors: | Shen, G, Rajan, R, Zhu, J, Bell, C.E, Pei, D. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and Synthesis of Substrate Analogue Inhibitors of S-Ribosylhomocysteinase (LuxS)
J.Med.Chem., 49, 2006
|
|
5J97
 
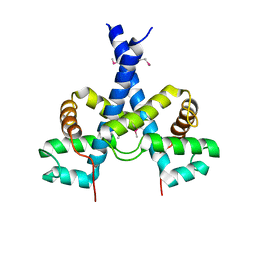 | |
3KO1
 
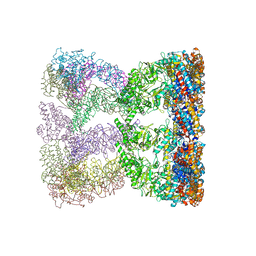 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
4RKA
 
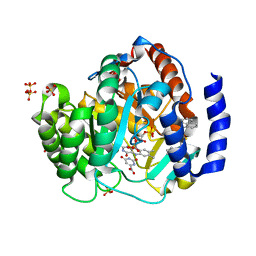 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347 | | Descriptor: | 2-{[5-(naphthalen-1-ylmethyl)-4-oxo-4H-1lambda~4~,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347
TO BE PUBLISHED
|
|
4RLI
 
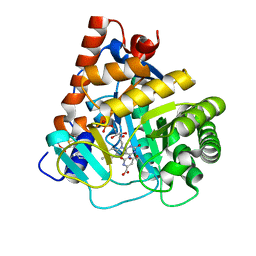 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048 | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048
TO BE PUBLISHED
|
|
4RK8
 
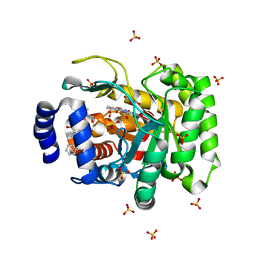 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356 | | Descriptor: | 5-fluoro-2-{[(5Z)-5-(naphthalen-1-ylmethylidene)-4-oxo-4,5-dihydro-1,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356
TO BE PUBLISHED
|
|
4V9N
 
 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
4WBE
 
 | |
4WBP
 
 | |
6KOB
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4UQ8
 
 | | Electron cryo-microscopy of bovine Complex I | | Descriptor: | ACYL CARRIER PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2014-06-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Architecture of Mammalian Respiratory Complex I.
Nature, 515, 2014
|
|
7U9F
 
 | | Integrin alaphIIBbeta3 complex with BMS compound 4 in Mn2+ | | Descriptor: | (4-{[(5S)-3-{4-[(E)-imino(4-methylpiperazin-1-yl)methyl]phenyl}-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.70000529 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UDG
 
 | | Integrin alaphIIBbeta3 complex with lotrafiban | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.800069 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UDH
 
 | | Integrin alaphIIBbeta3 complex with BMS4-3 | | Descriptor: | (4-{[(5S)-3-(piperidin-4-yl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99998844 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UK9
 
 | | Integrin alaphIIBbeta3 complex with lamifiban (Mn) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.60001969 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UH8
 
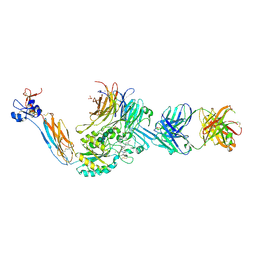 | | Integrin alaphIIBbeta3 complex with roxifiban (Mn/Ca) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.750024 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UFH
 
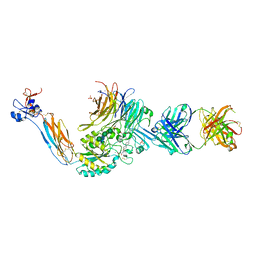 | | Integrin alaphIIBbeta3 complex with fradafiban (Mn/Ca) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99768162 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
