8D7E
 
 | |
8D6A
 
 | |
8D85
 
 | |
8D7H
 
 | |
2K73
 
 | | Solution NMR structure of integral membrane protein DsbB | | 分子名称: | Disulfide bond formation protein B | | 著者 | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | 登録日 | 2008-08-01 | | 公開日 | 2008-10-07 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K74
 
 | | Solution NMR structure of DsbB-ubiquinone complex | | 分子名称: | Disulfide bond formation protein B, UBIQUINONE-2 | | 著者 | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | 登録日 | 2008-08-01 | | 公開日 | 2008-10-07 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
6IXJ
 
 | | The crystal structure of sulfoacetaldehyde reductase from Klebsiella oxytoca | | 分子名称: | 2-hydroxyethylsulfonic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sulfoacetaldehyde reductase | | 著者 | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | 登録日 | 2018-12-10 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Biochemical and structural investigation of sulfoacetaldehyde reductase fromKlebsiella oxytoca.
Biochem. J., 476, 2019
|
|
6JKP
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense in complex with NAD+ | | 分子名称: | Methanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.008 Å) | | 主引用文献 | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6JKO
 
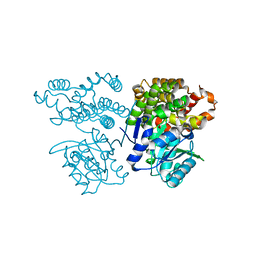 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense | | 分子名称: | Methanol dehydrogenase, ZINC ION | | 著者 | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | 登録日 | 2019-03-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6IV7
 
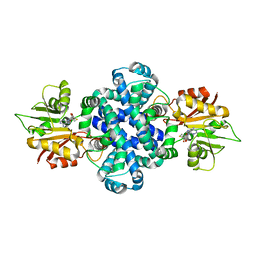 | | The crystal structure of a SAM-dependent enzyme from aspergillus flavus | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, methyltransferase lepI | | 著者 | Zhou, Y.Z, Peng, T, Liao, L.J, Liu, X.K, Zhao, Y.C, Guo, Y, Zeng, Z.X. | | 登録日 | 2018-12-02 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.937 Å) | | 主引用文献 | The crystal structure of A SAM-Dependent enzyme from Aspergillus flavus
To Be Published
|
|
6K5S
 
 | |
7DAJ
 
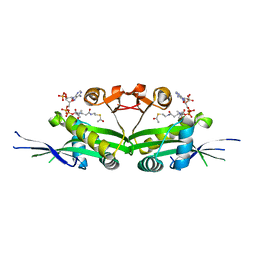 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | 分子名称: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | 分子名称: | Serotonin N-acetyltransferase 1, chloroplastic | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAL
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | 分子名称: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | 分子名称: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6IQL
 
 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | 分子名称: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | 著者 | Zhou, Y, Cao, C, Zhang, X.C. | | 登録日 | 2018-11-08 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
6KIM
 
 | |
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2023-03-06 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
5XOH
 
 | |
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | 分子名称: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | 著者 | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | 登録日 | 2002-07-25 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | 分子名称: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | 著者 | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | 登録日 | 2002-07-25 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | 分子名称: | Major vault protein | | 著者 | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | 登録日 | 2008-10-24 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | 分子名称: | Regulatory protein NPR1, ZINC ION | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6X3I
 
 | | NNAS Fc mutant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose | | 著者 | Wei, R, Zhou, Y.F. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.268 Å) | | 主引用文献 | Engineered Fc-glycosylation switch to eliminate antibody effector function.
Mabs, 12, 2020
|
|
5N15
 
 | | First Bromodomain (BD1) from Candida albicans Bdf1 in the unbound form | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing factor 1, GLYCEROL, ... | | 著者 | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | 登録日 | 2017-02-05 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
