1YGH
 
 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | 分子名称: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | 著者 | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | 登録日 | 1999-05-27 | | 公開日 | 1999-08-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5KPO
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | 分子名称: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | 登録日 | 2016-07-05 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KPQ
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | 分子名称: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | 登録日 | 2016-07-05 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
3HJW
 
 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | 分子名称: | 5'-R(*GP*AP*GP*CP*GP*(FHU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, POTASSIUM ION, ... | | 著者 | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
1ZLM
 
 | | Crystal structure of the SH3 domain of human osteoclast stimulating factor | | 分子名称: | Osteoclast stimulating factor 1 | | 著者 | Chen, L, Wang, Y, Wells, D, Toh, D, Harold, H, Zhou, J, DiGiammarino, E, Meehan, E.J. | | 登録日 | 2005-05-06 | | 公開日 | 2006-05-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Structure of the SH3 domain of human osteoclast-stimulating factor at atomic resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5KPN
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | 分子名称: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-propyl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | 登録日 | 2016-07-05 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
4IO0
 
 | | Crystal structure of F128A mutant of an epoxide hydrolase from Bacillus megaterium complexed with its product (R)-3-[1]naphthyloxy-propane-1,2-diol | | 分子名称: | (2R)-3-(naphthalen-1-yloxy)propane-1,2-diol, SULFATE ION, Soluble epoxide hydrolase | | 著者 | Kong, X.D, Zhou, J.H, Xu, J.H. | | 登録日 | 2013-01-07 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4INZ
 
 | | The crystal structure of M145A mutant of an epoxide hydrolase from Bacillus megaterium | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Soluble epoxide hydrolase | | 著者 | Kong, X.D, Zhou, J.H, Xu, J.H. | | 登録日 | 2013-01-07 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4HOW
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | 分子名称: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | 著者 | Xu, Z, Li, S, Xu, H, Zhou, J. | | 登録日 | 2012-10-22 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HPH
 
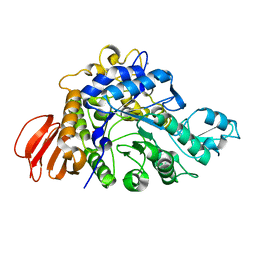 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | 分子名称: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | 著者 | Xu, Z, Li, S, Xu, H, Zhou, J. | | 登録日 | 2012-10-23 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOZ
 
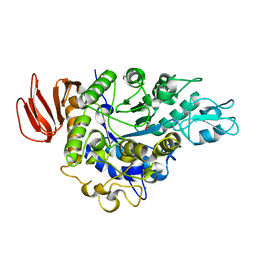 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | 分子名称: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | 著者 | Xu, Z, Li, S, Xu, H, Zhou, J. | | 登録日 | 2012-10-23 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | 分子名称: | short-chain reductase family member (5L265) | | 著者 | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-03-17 | | 公開日 | 2004-03-23 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1QSR
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | 分子名称: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | 著者 | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | 登録日 | 1999-06-23 | | 公開日 | 1999-09-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QST
 
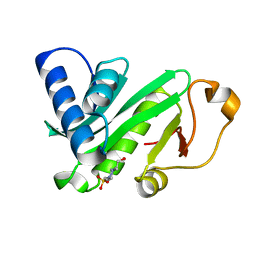 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | 著者 | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | 登録日 | 1999-06-23 | | 公開日 | 1999-09-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QSN
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | 分子名称: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | 著者 | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | 登録日 | 1999-06-22 | | 公開日 | 1999-09-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
3R1V
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, in complex with an azo compound | | 分子名称: | 4-{(E)-[4-(propan-2-yl)phenyl]diazenyl}phenol, Odorant binding protein, antennal | | 著者 | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | 登録日 | 2011-03-11 | | 公開日 | 2011-10-19 | | 最終更新日 | 2011-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1O
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges | | 分子名称: | Odorant binding protein, antennal, PALMITIC ACID | | 著者 | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | 登録日 | 2011-03-11 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1P
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, form P1 | | 分子名称: | Odorant binding protein, antennal, PALMITIC ACID | | 著者 | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | 登録日 | 2011-03-11 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3HJY
 
 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | 分子名称: | 5'-R(*GP*GP*AP*GP*CP*GP*UP*GP*CP*GP*GP*UP*UP*U)-3', 5'-R(*GP*GP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*CP*CP*GP*CP*GP*GP*CP*GP*C)-3', RNA (25-MER), ... | | 著者 | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
3R4S
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | 分子名称: | Botulinum neurotoxin type C1, N-acetyl-alpha-neuraminic acid, N-acetyl-beta-neuraminic acid | | 著者 | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | 登録日 | 2011-03-17 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
1CW3
 
 | | HUMAN MITOCHONDRIAL ALDEHYDE DEHYDROGENASE COMPLEXED WITH NAD+ AND MN2+ | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, MITOCHONDRIAL ALDEHYDE DEHYDROGENASE, ... | | 著者 | Ni, L, Zhou, J, Hurley, T.D, Weiner, H. | | 登録日 | 1999-08-25 | | 公開日 | 1999-08-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms.
Protein Sci., 8, 1999
|
|
3R4U
 
 | | Cell entry of botulinum neurotoxin type C is dependent upon interaction with two ganglioside molecules | | 分子名称: | Botulinum neurotoxin type C1 | | 著者 | Strotmeier, J, Gu, S, Jutzi, S, Mahrhold, S, Zhou, J, Pich, A, Bigalke, H, Rummel, A, Jin, R, Binz, T. | | 登録日 | 2011-03-17 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites.
Mol.Microbiol., 81, 2011
|
|
3V0C
 
 | | 4.3 angstrom crystal structure of an inactive BoNT/A (E224Q/R363A/Y366F) | | 分子名称: | BoNT/A, ZINC ION | | 著者 | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | 登録日 | 2011-12-07 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
7SCP
 
 | | The crystal structure of ScoE in complex with intermediate | | 分子名称: | (3R)-3-(oxaloamino)butanoic acid, 1,2-ETHANEDIOL, FE (II) ION, ... | | 著者 | Cha, L, Chen, J, Zhou, J, Chang, W. | | 登録日 | 2021-09-28 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Deciphering the Reaction Pathway of Mononuclear Iron Enzyme-Catalyzed N-C Triple Bond Formation in Isocyanide Lipopeptide and Polyketide Biosynthesis
Acs Catalysis, 12, 2022
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | 分子名称: | AvrPiz-t protein | | 著者 | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | 登録日 | 2012-07-23 | | 公開日 | 2012-09-12 | | 最終更新日 | 2013-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
