4E0H
 
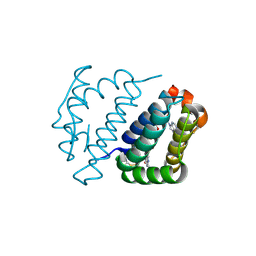 | | Crystal structure of FAD binding domain of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
2M80
 
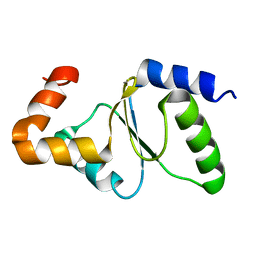 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
2P4Q
 
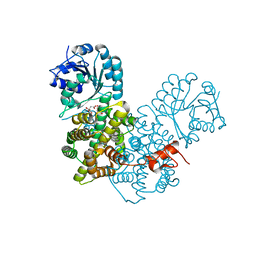 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
3IBH
 
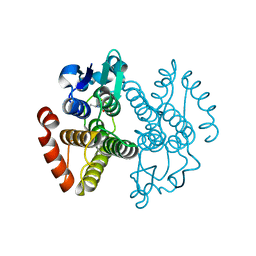 | | Crystal structure of Saccharomyces cerevisiae Gtt2 in complex with glutathione | | Descriptor: | GLUTATHIONE, Saccharomyces cerevisiae Gtt2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Bao, R, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2009-07-15 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
2BKW
 
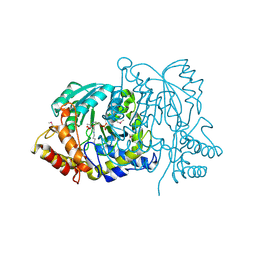 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
2FA4
 
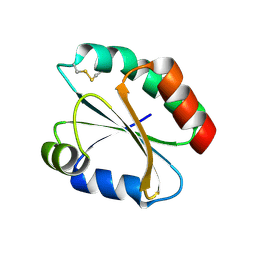 | |
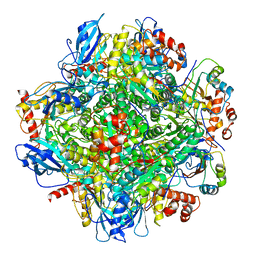
6LRR
 
 | | Cryo-EM structure of RuBisCO-Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LRS
 
 | | Cryo-EM structure of RbcL8-RbcS4 from Anabaena sp. PCC 7120 | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Sun, H, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LR0
 
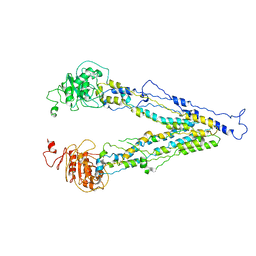 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
3LA7
 
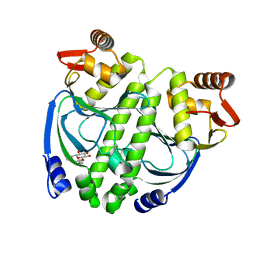 | | Crystal structure of NtcA in apo-form | | Descriptor: | Global nitrogen regulator, octyl beta-D-glucopyranoside | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Zhang, C.C, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ERF
 
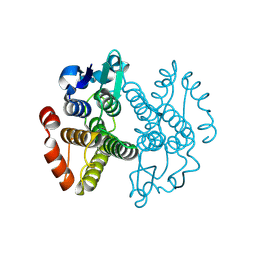 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae | | Descriptor: | Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
3LA2
 
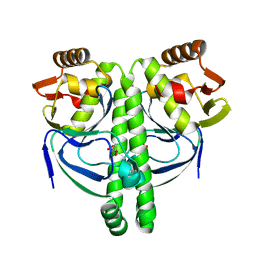 | | Crystal structure of NtcA in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LA3
 
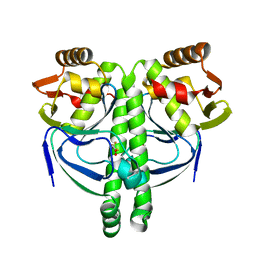 | | Crystal structure of NtcA in complex with 2,2-difluoropentanedioic acid | | Descriptor: | 2,2-difluoropentanedioic acid, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3EYX
 
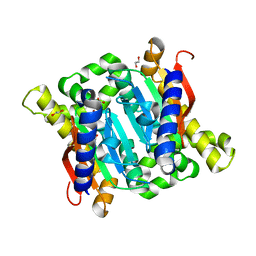 | | Crystal structure of Carbonic Anhydrase Nce103 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase, ... | | Authors: | Teng, Y.B, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2008-10-22 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the substrate tunnel of Saccharomyces cerevisiae carbonic anhydrase Nce103.
Bmc Struct.Biol., 9, 2009
|
|
3CMI
 
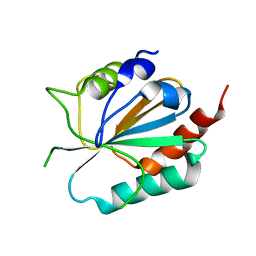 | |
3F3R
 
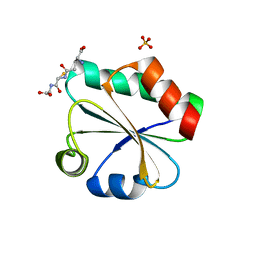 | | Crystal structure of yeast Thioredoxin1-glutathione mixed disulfide complex | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-1 | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3F3Q
 
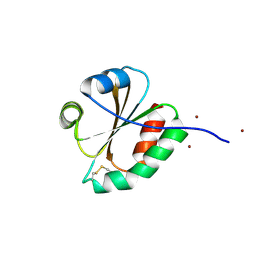 | | Crystal structure of the oxidised form of thioredoxin 1 from saccharomyces cerevisiae | | Descriptor: | Thioredoxin-1, ZINC ION | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3CTF
 
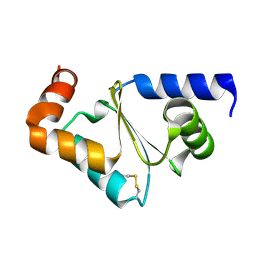 | | Crystal structure of oxidized GRX2 | | Descriptor: | Glutaredoxin-2 | | Authors: | Yu, J, Teng, Y.B, Zhou, C.Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the different activities of yeast Grx1 and Grx2.
Biochim.Biophys.Acta, 1804, 2010
|
|
3ERG
 
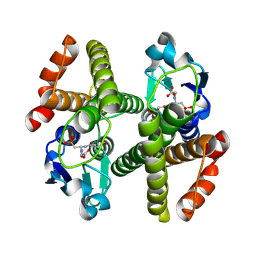 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae in complex with glutathione sulfnate | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
3CTG
 
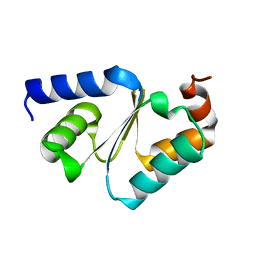 | |
4HRI
 
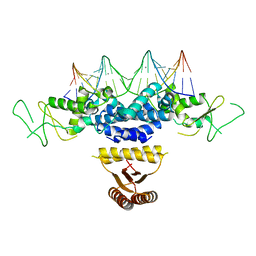 | | Crystal structure of HetR in complex with a 21-bp palindromic DNA at the upstream of the hetP promoter from Anabaena | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Chen, Y, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2012-10-28 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural and biochemical analyses of Anabaena HetR reveal insights into its binding to DNA targets and the inhibitory hexapeptide ERGSGR
To be Published
|
|
4I5V
 
 | | Crystal structure of yeast Ap4A phosphorylase Apa2 in complex with Ap4A | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2, BIS(ADENOSINE)-5'-TETRAPHOSPHATE | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4I5T
 
 | | Crystal structure of yeast Ap4A phosphorylase Apa2 | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2 | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
4IPN
 
 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
