6AD0
 
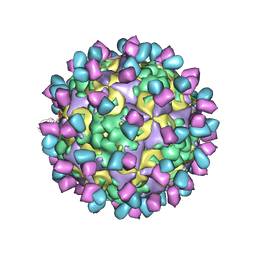 | | The structure of CVA10 mature virion in complex with Fab 2G8 | | Descriptor: | SPHINGOSINE, VH of Fab 2G8, VL of Fab 2G8, ... | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6ACZ
 
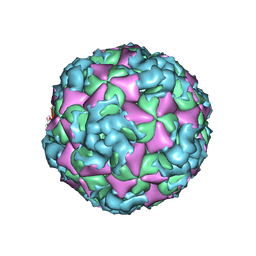 | | The structure of CVA10 virus A-particle from its complex with Fab 2G8 | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6ACY
 
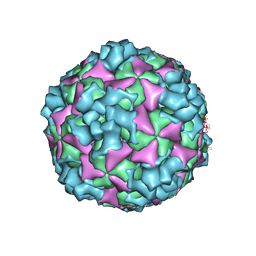 | | The structure of CVA10 virus A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Cui, Y.X, Zheng, Q.B, Zhu, R, Xu, L.F, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
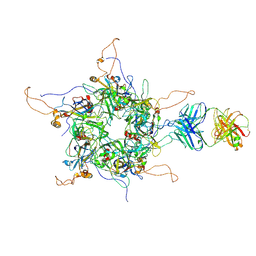 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
8X99
 
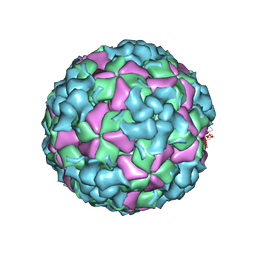 | | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2
To Be Published
|
|
8X98
 
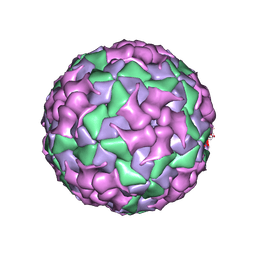 | | Cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8X95
 
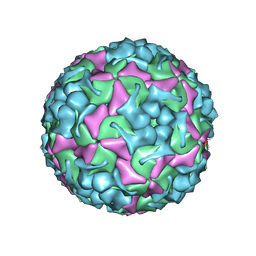 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8X9A
 
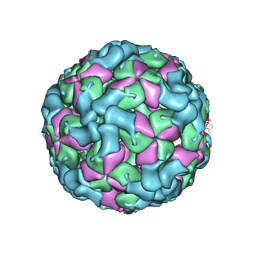 | | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2
To Be Published
|
|
8X96
 
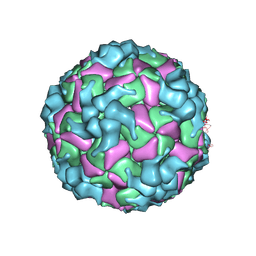 | | Cryo-EM structure of enterovirus A71 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of enterovirus A71 A-particle in complex with Fab h1A6.2
To Be Published
|
|
8X97
 
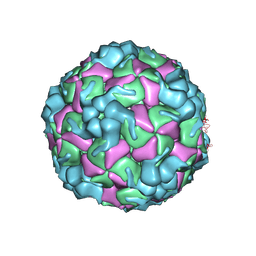 | | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2
To Be Published
|
|
8XEW
 
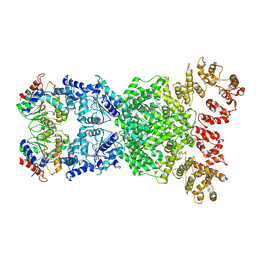 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein | | Descriptor: | DSR2 H171A | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
8XFF
 
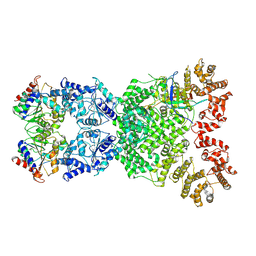 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein in complex with SPR phage tail tube protein | | Descriptor: | DSR2(H171A), SPR tail tube protein | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
8XFE
 
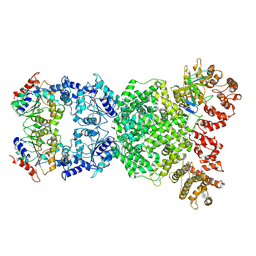 | | Cryo-EM structure of defence-associated sirtuin 2 (DSR2) H171A protein in complex with DSR anti-defence 1(DSAD1) | | Descriptor: | DSAD1, DSR2(H171A) | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
2GUV
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
2GUS
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
