2OF6
 
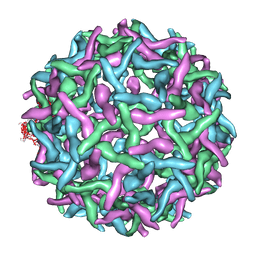 | | Structure of immature West Nile virus | | Descriptor: | envelope glycoprotein E | | Authors: | Zhang, Y, Kaufmann, B, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-01-02 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structure of immature west nile virus.
J.Virol., 81, 2007
|
|
2PQN
 
 | |
2PQR
 
 | |
3TG3
 
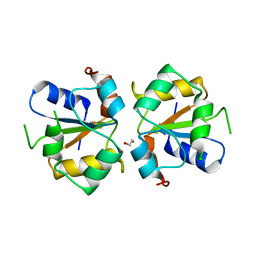 | | Crystal structure of the MAPK binding domain of MKP7 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | Authors: | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
8HLD
 
 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
8HLC
 
 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
3TG1
 
 | |
3RY0
 
 | | Crystal structure of TomN, a 4-Oxalocrotonate Tautomerase homologue in Tomaymycin biosynthetic pathway | | Descriptor: | Putative tautomerase | | Authors: | Zhang, Y, Yan, W.P, Li, W.Z, Whitman, C.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kinetic, Crystallographic, and Mechanistic Characterization of TomN: Elucidation of a Function for a 4-Oxalocrotonate Tautomerase Homologue in the Tomaymycin Biosynthetic Pathway.
Biochemistry, 50, 2011
|
|
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
7WFY
 
 | |
6JJE
 
 | |
6JJG
 
 | |
6KFL
 
 | | Crystal structure of a two-quartet DNA G-quadruplex complexed with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), ... | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a two-quartet DNA G-quadruplex complexed with the porphyrin TMPyP4
To Be Published
|
|
3UUX
 
 | | Crystal structure of yeast Fis1 in complex with Mdv1 fragment containing N-terminal extension and coiled coil domains | | Descriptor: | Mitochondria fission 1 protein, Mitochondrial division protein 1 | | Authors: | Zhang, Y, Chan, N.C, Gristick, H, Chan, D.C. | | Deposit date: | 2011-11-28 | | Release date: | 2012-02-08 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (mdv1) coiled coil.
J.Biol.Chem., 287, 2012
|
|
7W7V
 
 | | 'late' E2P of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7U
 
 | | The 'Ca2+-unbound' BeF3- of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7W
 
 | | E2 Pi of SERCA2b | | Descriptor: | MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2, TETRAFLUOROALUMINATE ION | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7T
 
 | | The E1-BeF3- 2Ca2+ of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
7EN5
 
 | | The crystal structure of Escherichia coli MurR in complex with N-acetylglucosamine-6-phosphate | | Descriptor: | 2-METHOXYETHANOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN7
 
 | | The crystal structure of Escherichia coli MurR in complex with N-acetylmuramic-acid-6-phosphate | | Descriptor: | (2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2,5-bis(oxidanyl)-6-(phosphonooxymethyl)oxan-4-yl]oxypropanoic acid, HTH-type transcriptional regulator MurR | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7CZH
 
 | | PL24 ulvan lyase-Uly1 | | Descriptor: | CALCIUM ION, GLYCEROL, Uly1 | | Authors: | Zhang, Y.Z, Chen, X.L, Dong, F, Xu, F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Mechanistic Insights into Substrate Recognition and Catalysis of a New Ulvan Lyase of Polysaccharide Lyase Family 24.
Appl.Environ.Microbiol., 87, 2021
|
|
7DR8
 
 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
