3J9E
 
 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | 分子名称: | VP5 | | 著者 | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | 登録日 | 2015-01-10 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | 著者 | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
5K3H
 
 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | 分子名称: | Acyl-coenzyme A oxidase | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3I
 
 | | Crystal structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans complexed with FAD and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.683 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3G
 
 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | 分子名称: | Acyl-coenzyme A oxidase | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3J
 
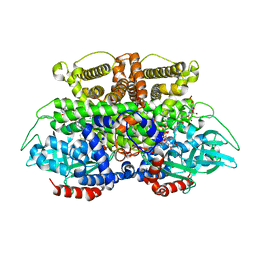 | | Crystals structure of Acyl-CoA oxidase-2 in Caenorhabditis elegans bound with FAD, ascaroside-CoA, and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | 登録日 | 2016-05-19 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5NWT
 
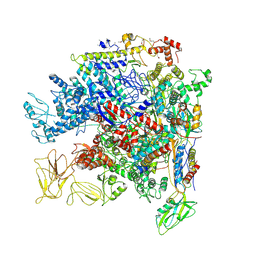 | | Crystal Structure of Escherichia coli RNA polymerase - Sigma54 Holoenzyme complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zhang, X, Buck, M, Darbari, V.C, Yang, Y, Zhang, N, Lu, D, Glyde, R, Wang, Y, Winkelman, J, Gourse, R.L, Murakami, K.S. | | 登録日 | 2017-05-08 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.76 Å) | | 主引用文献 | TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
5ZMD
 
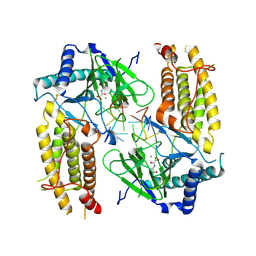 | | Crystal structure of FTO in complex with m6dA modified ssDNA | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase FTO, DNA (5'-D(P*TP*CP*TP*(6MA)P*TP*AP*TP*CP*G)-3'), MANGANESE (II) ION, ... | | 著者 | Zhang, X, Wei, L.H, Luo, J, Xiao, Y, Liu, J, Zhang, W, Zhang, L, Jia, G.F. | | 登録日 | 2018-04-02 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | 分子名称: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Zhang, X, Zhang, L, Zhang, S. | | 登録日 | 2018-08-17 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.78 Å) | | 主引用文献 | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
3IYL
 
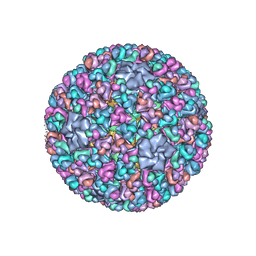 | | Atomic CryoEM Structure of a Nonenveloped Virus Suggests How Membrane Penetration Protein is Primed for Cell Entry | | 分子名称: | Core protein VP6, MYRISTIC ACID, Outer capsid VP4, ... | | 著者 | Zhang, X, Jin, L, Fang, Q, Hui, W, Zhou, Z.H. | | 登録日 | 2010-02-02 | | 公開日 | 2010-05-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry.
Cell(Cambridge,Mass.), 141, 2010
|
|
3J4U
 
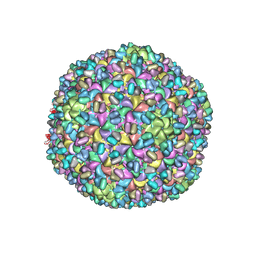 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | 分子名称: | cementing protein, major capsid protein | | 著者 | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | 登録日 | 2013-10-09 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
4DXW
 
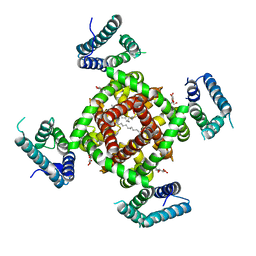 | | Crystal structure of NavRh, a voltage-gated sodium channel | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | 著者 | Zhang, X, Ren, W.L, Yan, C.Y, Wang, J.W, Yan, N. | | 登録日 | 2012-02-28 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.052 Å) | | 主引用文献 | Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel
Nature, 486, 2012
|
|
4N6E
 
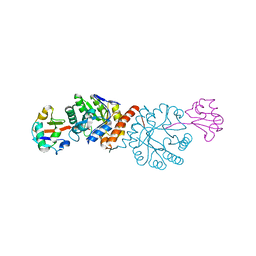 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | 分子名称: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | 著者 | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | 登録日 | 2013-10-11 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4N6F
 
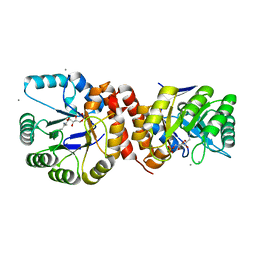 | | Crystal structure of Amycolatopsis orientalis BexX complexed with G6P | | 分子名称: | CALCIUM ION, FRUCTOSE -6-PHOSPHATE, Putative thiosugar synthase | | 著者 | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | 登録日 | 2013-10-11 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | 登録日 | 2010-04-14 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
4O9L
 
 | |
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
4O9F
 
 | |
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | 著者 | Zhang, X, Zhang, Q, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
5YB2
 
 | | Crystal structure of LP-11/N44 | | 分子名称: | Envelope glycoprotein, LP-11 | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YB4
 
 | | Crystal structure of HP23LN36KR | | 分子名称: | HP23L, N36KR | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YB3
 
 | | Crystal structure of HP23L/N36 | | 分子名称: | Envelope glycoprotein, HP23L | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-09-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.043 Å) | | 主引用文献 | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
4OA7
 
 | | Crystal structure of Tankyrase1 in complex with IWR1 | | 分子名称: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | 著者 | Zhang, X, He, H. | | 登録日 | 2014-01-03 | | 公開日 | 2015-01-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
