8YN7
 
 | | Cryo-EM structure of histamine H3 receptor in complex with immethridine and miniGo | | Descriptor: | 4-(1H-imidazol-4-ylmethyl)pyridine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN2
 
 | | Cryo-EM structure of histamine H1 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN9
 
 | | Cryo-EM structure of histamine H4 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN5
 
 | | Cryo-EM structure of histamine H3 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN4
 
 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN3
 
 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGs | | Descriptor: | CHOLESTEROL, Engineered guanine nucleotide,binding protein G(s) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YNA
 
 | | Cryo-EM structure of histamine H4 receptor in complex with immepip and Gi | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN6
 
 | | Cryo-EM structure of histamine H3 receptor in complex with imetit and Gi | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
3KLX
 
 | |
3JB7
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
5NWT
 
 | | Crystal Structure of Escherichia coli RNA polymerase - Sigma54 Holoenzyme complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhang, X, Buck, M, Darbari, V.C, Yang, Y, Zhang, N, Lu, D, Glyde, R, Wang, Y, Winkelman, J, Gourse, R.L, Murakami, K.S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
2Q12
 
 | | Crystal Structure of BAR domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhang, X.C, Zhu, G. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
1NUT
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUU
 
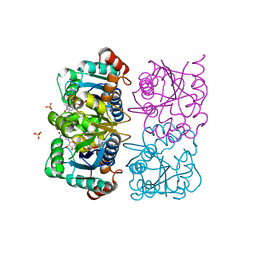 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NAD | | Descriptor: | FKSG76, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUS
 
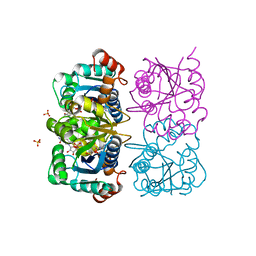 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG AND NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, ... | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUP
 
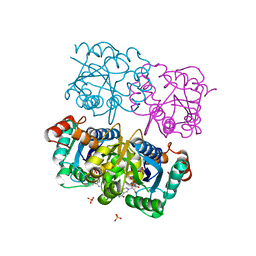 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEX WITH NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUQ
 
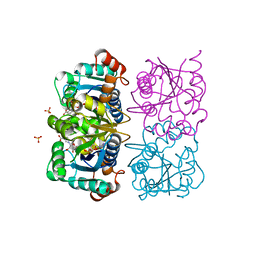 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NaAD | | Descriptor: | FKSG76, NICOTINIC ACID ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUR
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE | | Descriptor: | FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
8YN8
 
 | | Cryo-EM structure of histamine H3 receptor in complex with proxyfan and miniGo | | Descriptor: | 5-(3-phenylmethoxypropyl)-1H-imidazole, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
1DYD
 
 | |
1DYC
 
 | |
8T3O
 
 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
