8Z5D
 
 | |
8Z5F
 
 | |
8Z5C
 
 | |
6RKZ
 
 | | Recombinant Pseudomonas stutzeri nitrous oxide reductase, form II | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Wuest, A, Prasser, B, Mueller, C, Einsle, O. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Functional assembly of nitrous oxide reductase provides insights into copper site maturation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RL0
 
 | | Recombinant Pseudomonas stutzeri nitrous oxide reductase, form I | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Wuest, A, Prasser, B, Mueller, C, Einsle, O. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Functional assembly of nitrous oxide reductase provides insights into copper site maturation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8JNK
 
 | |
8JNR
 
 | |
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y7M
 
 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Hilgenfeld, R. | | Deposit date: | 2020-03-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2E
 
 | |
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
5H9G
 
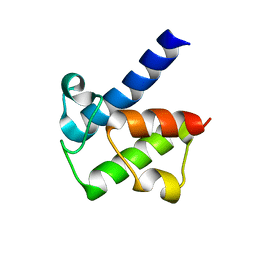 | |
5H9H
 
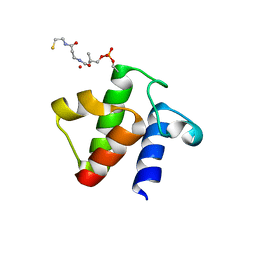 | |
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8JKK
 
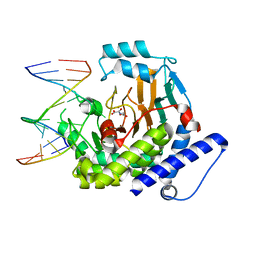 | |
6B8F
 
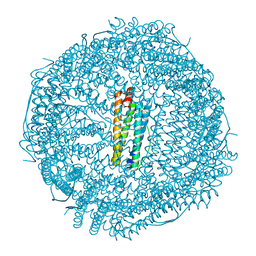 | | Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | Authors: | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | Deposit date: | 2017-10-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
6Y72
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H178A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y71
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H130A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y6Y
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H129A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7E
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H494A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7D
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H433A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
6Y77
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H326A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINUCLEAR COPPER ION, FORMIC ACID, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
7ZJE
 
 | | C16-2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2,Enhanced green fluorescent protein | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJD
 
 | |
