3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | 分子名称: | CAFFEINE, Chitinase | | 著者 | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | 登録日 | 2009-02-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
5XG5
 
 | | Crystal structure of Mitsuba-1 with bound NAcGal | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | 著者 | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | 登録日 | 2017-04-12 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
5EWP
 
 | | ARO (armadillo repeats only protein) from Plasmodium falciparum | | 分子名称: | ARO (armadillo repeats only protein) | | 著者 | Brown, C, Zhang, K, Emery, J, Prusty, D, Wetzel, J, Heincke, D, Gilberger, T, Junop, M. | | 登録日 | 2015-11-20 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | ARO (armadillo repeats only protein) from Plasmodium falciparum
To Be Published
|
|
4NPH
 
 | | Crystal structure of SsaN from Salmonella enterica | | 分子名称: | Probable secretion system apparatus ATP synthase SsaN | | 著者 | Sugiman-Marangos, S.N, Zhang, K, Cooper, C.A, Coombes, B.K, Junop, M.S. | | 登録日 | 2013-11-21 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of SsaN from Salmonella enterica
J.Biol.Chem., 2014
|
|
7XML
 
 | |
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | 分子名称: | MAGNESIUM ION, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.732 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
5HSJ
 
 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
7W16
 
 | | Complex structure of alginate lyase AlyV with M8 | | 分子名称: | GLYCEROL, alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-08-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W18
 
 | | Complex structure of alginate lyase PyAly with M5 | | 分子名称: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W13
 
 | | Complex structure of alginate lyase PyAly with M8 | | 分子名称: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W12
 
 | | Complex structure of alginate lyase AlyB-OU02 with G9 | | 分子名称: | Alginate lyase, SULFATE ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7YGA
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGB
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YG9
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing | | 分子名称: | MAGNESIUM ION, RNA (391-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | 著者 | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | 登録日 | 2022-07-11 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | 分子名称: | Motility protein FimV | | 著者 | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | 登録日 | 2013-08-19 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.006 Å) | | 主引用文献 | Crystal structure of TPR2 from FimV
To be Published
|
|
3T9K
 
 | | Crystal Structure of ACAP1 C-portion mutant S554D fused with integrin beta1 peptide | | 分子名称: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1,Peptide from Integrin beta-1, SULFATE ION, ... | | 著者 | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | 登録日 | 2011-08-03 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
6BD0
 
 | | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex) | | 分子名称: | CALCIUM ION, DNA (25-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | 著者 | Brown, C, Zhang, K, Laforet, M, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex)
To Be Published
|
|
6BDB
 
 | | I-OnuI K227Y, D236A bound to A3G substrate (pre-cleavage complex) | | 分子名称: | DNA (26-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | 著者 | Brown, C, Zhang, K, McMurrough, T.A, Laforet, M, Gloor, G.B, Edgell, D.R, Junop, M. | | 登録日 | 2017-10-22 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | I-OnuI K227Y, D236A bound to A3G substrate (pre-cleavage complex)
To Be Published
|
|
6BCT
 
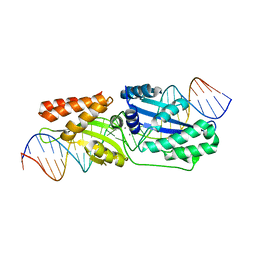 | | I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex) | | 分子名称: | CALCIUM ION, DNA (26-MER), DNA (27-MER), ... | | 著者 | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
7D3U
 
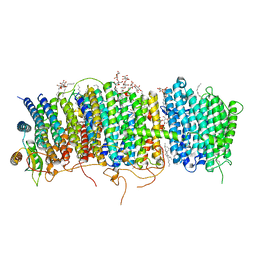 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | 分子名称: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | 著者 | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | 登録日 | 2020-09-21 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6BCE
 
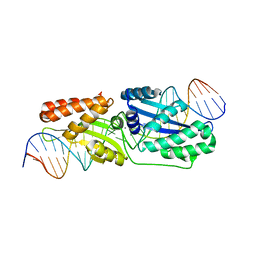 | | Wild-type I-LtrI bound to cognate substrate (pre-cleavage complex) | | 分子名称: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | 著者 | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
6BCI
 
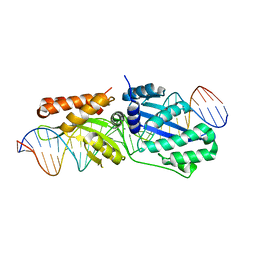 | | Wild-type I-LtrI bound to non-cognate C4 substrate (pre-cleavage complex) | | 分子名称: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | 著者 | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
3U9G
 
 | | Crystal structure of the Zinc finger antiviral protein | | 分子名称: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | 著者 | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | 登録日 | 2011-10-18 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3J17
 
 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | 分子名称: | Structural protein VP3, Structural protein VP5, VP1 | | 著者 | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | 登録日 | 2011-12-25 | | 公開日 | 2012-04-04 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3K6J
 
 | | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans | | 分子名称: | PHOSPHATE ION, Protein F01G10.3, confirmed by transcript evidence, ... | | 著者 | Ouyang, Z, Zhang, K, Zhai, Y, Lu, J, Sun, F. | | 登録日 | 2009-10-09 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the dehydrogenase part of multifuctional enzyme 1 from C.elegans
To be Published
|
|
