5ZY9
 
 | | Structural basis for a tRNA synthetase | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonyl-tRNA synthase, ZINC ION | | Authors: | Zhang, J. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for inhibition of
translational functions on a tRNA synthetase
To Be Published
|
|
8XAI
 
 | |
7M5D
 
 | |
7N98
 
 | | Cryo-EM structure of MFSD2A | | Descriptor: | Sodium-dependent lysophosphatidylcholine symporter 1 | | Authors: | Zhang, J, Feng, L. | | Deposit date: | 2021-06-17 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of blood-brain-barrier lipid transporter MFSD2A.
Nature, 596, 2021
|
|
8W8D
 
 | |
8TDM
 
 | | Cryo-EM structure of AtMSL10-K539E | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDJ
 
 | |
8TDL
 
 | |
8TDK
 
 | | Cryo-EM structure of AtMSL10-G556V | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
2M8V
 
 | | Solution Structure and Activity Study of Bovicin HJ50, a Particular Type AII Lantibiotic | | Descriptor: | BovA | | Authors: | Zhang, J, Feng, Y, Wang, J, Zhong, J. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Type AII lantibiotic bovicin HJ50 with a rare disulfide bond: structure, structure-activity relationships and mode of action.
Biochem.J., 461, 2014
|
|
2NWM
 
 | |
6LZ1
 
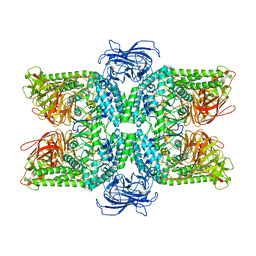 | | Structure of S.pombe alpha-mannosidase Ams1 | | Descriptor: | Ams1, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of fission yeast tetrameric alpha-mannosidase Ams1.
Febs Open Bio, 10, 2020
|
|
2GMH
 
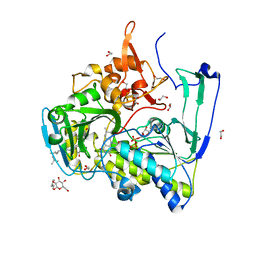 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase in Complexed with Ubiquinone | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, Electron transfer flavoprotein-ubiquinone oxidoreductase, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7YUK
 
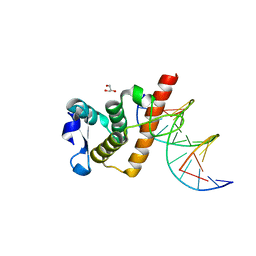 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
2GMJ
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase | | Descriptor: | Electron transfer flavoprotein-ubiquinone oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5H3V
 
 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2FGF
 
 | |
3UIT
 
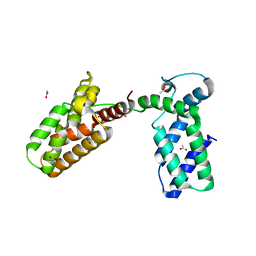 | | Overall structure of Patj/Pals1/Mals complex | | Descriptor: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | Authors: | Zhang, J, Yang, X, Long, J, Shen, Y. | | Deposit date: | 2011-11-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
7V5H
 
 | |
7V9Z
 
 | |
7VA3
 
 | |
7VA6
 
 | |
7VA2
 
 | |
7V3P
 
 | | Cryo-EM structure of the IGF1R/insulin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Zhang, J, Liu, C, Zhang, X, Wei, T, Wu, C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the IGF1R/insulin complex
To Be Published
|
|
