7ZJD
 
 | |
7ZJI
 
 | |
7ZJG
 
 | | Probenecid | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
7ZJH
 
 | | TRPV2-C16+Pro-2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zhang, L, Gourdon, P, Zygmunt, P.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cannabinoid non-cannabidiol site modulation of TRPV2 structure and function.
Nat Commun, 13, 2022
|
|
8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
8AHX
 
 | |
7AQA
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H382A | | Descriptor: | (dicuprio-$l^{3}-sulfanyl)-sulfanyl-copper, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
8C9S
 
 | |
6Y2E
 
 | |
7QUW
 
 | | CVB3-3Cpro in complex with inhibitor MG-78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
5MGY
 
 | | Crystal structure of Pseudomonas stutzeri flavinyl transferase ApbE, apo form | | Descriptor: | FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zhang, L, Trncik, C, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2016-11-22 | | Release date: | 2016-12-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flavinyl transferase ApbE of Pseudomonas stutzeri matures the NosR protein required for nitrous oxide reduction.
Biochim. Biophys. Acta, 1858, 2016
|
|
3CF8
 
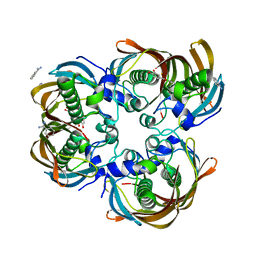 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with quercetin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, BENZAMIDINE, ... | | Authors: | Zhang, L, Wu, D, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2008-03-03 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3CF9
 
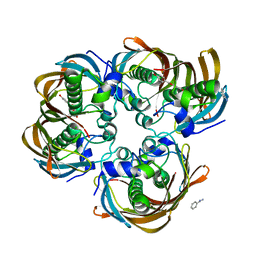 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with apigenin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, BENZAMIDINE, ... | | Authors: | Zhang, L, Wu, D, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2008-03-03 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
6Y6Y
 
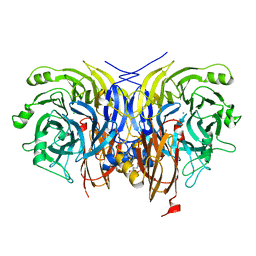 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H129A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7E
 
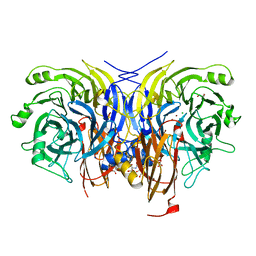 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H494A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7D
 
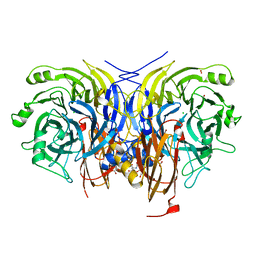 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H433A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
4ZJB
 
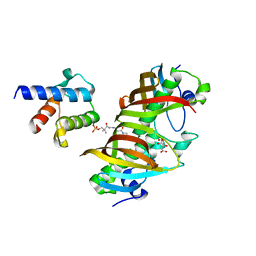 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Zhang, L, Zhang, L, Shen, X, Jiang, H. | | Deposit date: | 2015-04-29 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis.
Cell Res., 26, 2016
|
|
8B35
 
 | |
8B32
 
 | |
8B3C
 
 | | Chalcone synthase from Hordeum vulgare complexed with CoA and eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, Chalcone synthase 2 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Plant Polyketide Synthase for the Biosynthesis of Methylated Flavonoids.
J.Agric.Food Chem., 72, 2024
|
|
7AQ8
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y/D576A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ5
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583N | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ6
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583F | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ7
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583Y | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AQ3
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583D | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
