7XM9
 
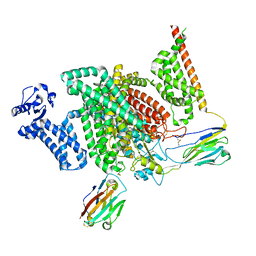 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6BWF
 
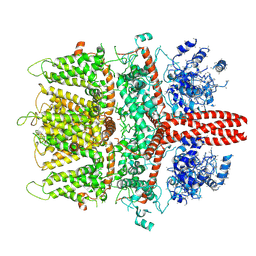 | | 4.1 angstrom Mg2+-unbound structure of mouse TRPM7 | | Descriptor: | TRPM7 | | Authors: | Zhang, J, Li, Z, Duan, J, Abiria, S.A, Clapham, D.E. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7KJ2
 
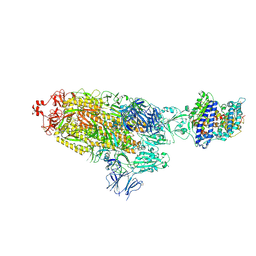 | | SARS-CoV-2 Spike Glycoprotein with one ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ4
 
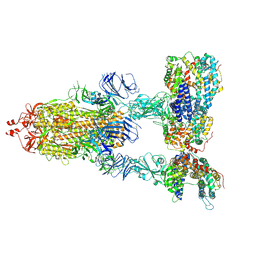 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3UIT
 
 | | Overall structure of Patj/Pals1/Mals complex | | Descriptor: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | Authors: | Zhang, J, Yang, X, Long, J, Shen, Y. | | Deposit date: | 2011-11-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
2NWM
 
 | |
8IW5
 
 | | Crystal structure of liprin-beta H2H3 dimer | | Descriptor: | CALCIUM ION, Liprin-beta-1 | | Authors: | Zhang, J, Chen, S, Wei, Z. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
8TDM
 
 | | Cryo-EM structure of AtMSL10-K539E | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDJ
 
 | |
8TDK
 
 | | Cryo-EM structure of AtMSL10-G556V | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDL
 
 | |
7XG3
 
 | | CryoEM structure of type IV-A CasDinG bound NTS-nicked Csf-crRNA-dsDNA quaternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG0
 
 | | CryoEM structure of type IV-A Csf-crRNA-dsDNA ternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG4
 
 | | CryoEM structure of type IV-A CasDinG bound NTS-nicked Csf-crRNA-dsDNA quaternary complex in a second state | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG2
 
 | | CryoEM structure of type IV-A NTS-nicked dsDNA bound Csf-crRNA ternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7YFM
 
 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
 | |
7YUK
 
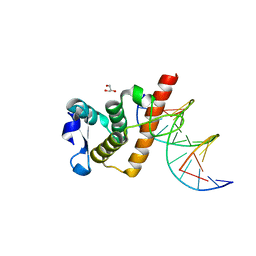 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
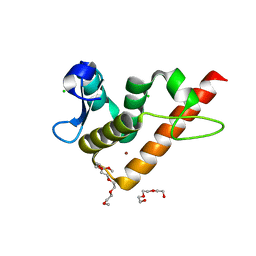 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7KRR
 
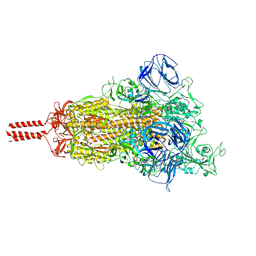 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRS
 
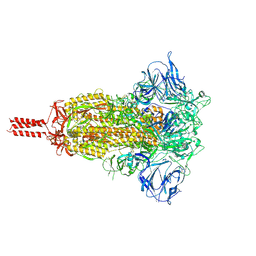 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRQ
 
 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
4G3H
 
 | | Crystal structure of helicobacter pylori arginase | | Descriptor: | Arginase (RocF), MANGANESE (II) ION | | Authors: | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
3ST9
 
 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
