4C7L
 
 | | Crystal structure of Mouse Hepatitis virus strain S Hemagglutinin- esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Huizinga, E.G. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Murine Coronavirus Hemagglutinin-Esterase Receptor-Binding Site: A Major Shift in Ligand Specificity Through Modest Changes in Architecture.
Plos Pathog., 8, 2012
|
|
4C7W
 
 | |
5JIF
 
 | | Crystal structure of mouse hepatitis virus strain DVIM Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2016-04-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3I1L
 
 | |
3I27
 
 | | Structure of bovine torovirus Hemagglutinin-Esterase in complex with receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Huizinga, E.G. | | Deposit date: | 2009-06-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I26
 
 | | Structure of bovine torovirus Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Huizinga, E.G. | | Deposit date: | 2009-06-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I1K
 
 | | Structure of porcine torovirus Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Hemagglutinin-esterase protein | | Authors: | Zeng, Q.H, Huizinga, E.G. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3CL4
 
 | | Crystal structure of bovine coronavirus hemagglutinin-esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CL5
 
 | | Structure of coronavirus hemagglutinin-esterase in complex with 4,9-O-diacetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4MZ4
 
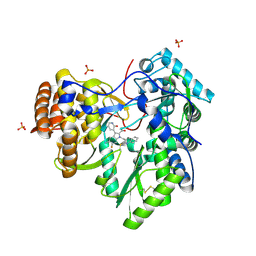 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5J5N
 
 | |
8WXB
 
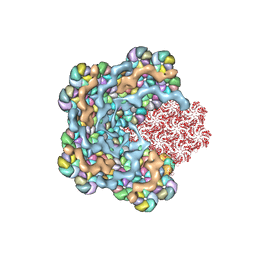 | | Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4 | | Descriptor: | Carboxysome assembly protein CsoS2, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1 | | Authors: | Jiang, Y.L, Zhou, R.Q, Zhou, C.Z, Zeng, Q.L. | | Deposit date: | 2023-10-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and assembly of the alpha-carboxysome in the marine cyanobacterium Prochlorococcus.
Nat.Plants, 10, 2024
|
|
2YOJ
 
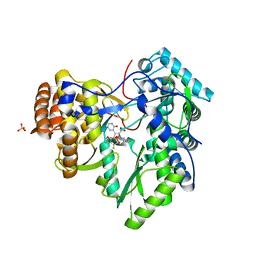 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
7Y55
 
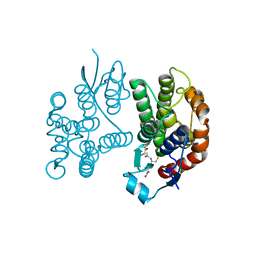 | |
5J4U
 
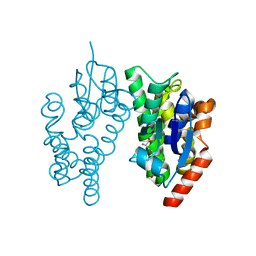 | |
1J1L
 
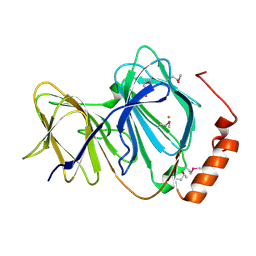 | | Crystal structure of human Pirin: a Bcl-3 and Nuclear factor I interacting protein and a cupin superfamily member | | Descriptor: | FE (II) ION, Pirin | | Authors: | Pang, H, Bartlam, M, Zeng, Q, Gao, G.F, Rao, Z. | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor
J.Biol.Chem., 279, 2004
|
|
1PFT
 
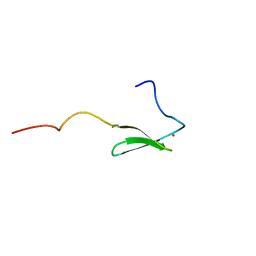 | | N-TERMINAL DOMAIN OF TFIIB, NMR | | Descriptor: | TFIIB, ZINC ION | | Authors: | Zhu, W, Zeng, Q, Colangelo, C.M, Lewis, L.M, Summers, M.F, Scott, R.A. | | Deposit date: | 1996-03-27 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon.
Nat.Struct.Biol., 3, 1996
|
|
3U4R
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3U4O
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7DXM
 
 | | Crystal structure of DltD | | Descriptor: | Protein DltD, SULFATE ION | | Authors: | Yan, X.X, Zeng, Q, Tian, L.F. | | Deposit date: | 2021-01-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of an O-acyltransfer Terminal Protein stDltD and Its Implications for dlt Operon-mediated D-alanylation of S. thermophilus.
Prog.Biochem.Biophys., 48, 2022
|
|
