3LGD
 
 | |
3LGG
 
 | | Crystal structure of human adenosine deaminase growth factor, adenosine deaminase type 2 (ADA2) complexed with transition state analogue, coformycin | | Descriptor: | (8R)-3-beta-D-ribofuranosyl-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Adenosine deaminase CECR1, ... | | Authors: | Zavialov, A.V. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the growth factor activity of human adenosine deaminase ADA2.
J.Biol.Chem., 285, 2010
|
|
1P5V
 
 | | X-ray structure of the Caf1M:Caf1 chaperone:subunit preassembly complex | | Descriptor: | Chaperone protein Caf1M, F1 capsule antigen | | Authors: | Zavialov, A.V, Berglund, J, Pudney, A.F, Fooks, L.J, Ibrahim, T.M, MacIntyre, S, Knight, S.D. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Biogenesis of the Capsular F1 Antigen from Yersinia pestis. Preserved Folding Energy Drives Fiber Formation
Cell(Cambridge,Mass.), 113, 2003
|
|
1P5U
 
 | | X-ray structure of the ternary Caf1M:Caf1:Caf1 chaperone:subunit:subunit complex | | Descriptor: | Chaperone protein Caf1M, F1 capsule antigen | | Authors: | Zavialov, A.V, Berglund, J, Pudney, A.F, Fooks, L.J, Ibrahim, T.M, MacIntyre, S, Knight, S.D. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and Biogenesis of the Capsular F1 Antigen from Yersinia pestis. Preserved Folding Energy Drives Fiber Formation
Cell(Cambridge,Mass.), 113, 2003
|
|
1Z9S
 
 | | Crystal Structure of the native chaperone:subunit:subunit Caf1M:Caf1:Caf1 complex | | Descriptor: | Chaperone protein Caf1M, F1 capsule antigen | | Authors: | Zavialov, A.V, Tischenko, V.M, Fooks, L.J, Brandsdal, B.O, Aqvist, J, Zav'yalov, V.P, Macintyre, S, Knight, S.D. | | Deposit date: | 2005-04-04 | | Release date: | 2005-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resolving the energy paradox of chaperone/usher-mediated fibre assembly
Biochem.J., 389, 2005
|
|
7ZL4
 
 | | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | | Descriptor: | CsuA/B | | Authors: | Pakharukova, N, Malmi, H, Tuittila, M, Paavilainen, S, Ghosal, D, Chang, Y.W, Jensen, G.J, Zavialov, A.V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.
Nature, 609, 2022
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
6FQA
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Parilova, O, Pakharukova, N.A, Malmi, H, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
1MI6
 
 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
6FJY
 
 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1ML5
 
 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | Deposit date: | 2002-08-30 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
4PH8
 
 | |
1ZN1
 
 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1ZN0
 
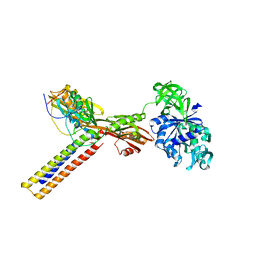 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | Descriptor: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
6FM5
 
 | | Crystal structure of self-complemented CsuA/B major subunit from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B,CsuA/B,CsuA/B | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6FQ0
 
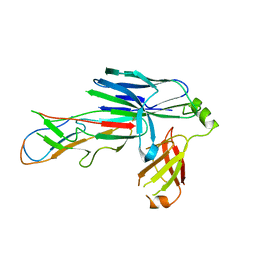 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
2MPV
 
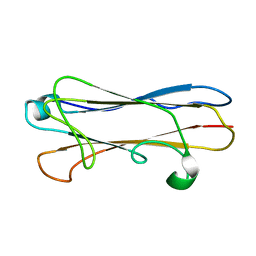 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
4AZ8
 
 | | Crystal structure of the complex of the Caf1M:Caf1 chaperone:subunit preassembly complex carrying the KDKDTN insertion at the F1G1 loop region | | Descriptor: | CHAPERONE PROTEIN CAF1M, F1 CAPSULE ANTIGEN | | Authors: | Yu, X.D, Dubnovitsky, A, Pudney, A.F, MacIntyre, S, Knight, S.D, Zavialov, A.V. | | Deposit date: | 2012-06-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Allosteric Mechanism Controls Traffic in the Chaperone/Usher Pathway.
Structure, 20, 2012
|
|
4AY0
 
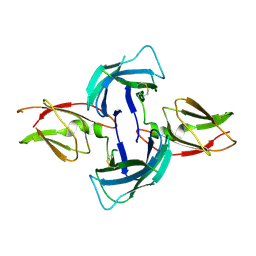 | | High resolution crystal structure of the monomeric subunit-free Caf1M chaperone | | Descriptor: | CHAPERONE PROTEIN CAF1M | | Authors: | Yu, X.D, Dubnovitsky, A, Pudney, A.F, MacIntyre, S, Knight, S.D, Zavialov, A.V. | | Deposit date: | 2012-06-16 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Allosteric Mechanism Controls Traffic in the Chaperone/Usher Pathway.
Structure, 20, 2012
|
|
4B9G
 
 | | Structure of CssB subunit complemented with donor strand from CssA subunit of enterotoxigenic Escherichia coli colonization factor CS6 | | Descriptor: | CS6 FIMBRIAL SUBUNIT B, CS6 FIMBRIAL SUBUNIT A | | Authors: | Roy, S.P, Rahman, M.M, Yu, X.D, Tuittila, M, Knight, S.D, Zavialov, A.V. | | Deposit date: | 2012-09-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of Enterotoxigenic Escherichia Coli Colonization Factor Cs6 Reveals a Novel Type of Functional Assembly.
Mol.Microbiol., 86, 2012
|
|
5LN8
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica, in complex with galactose | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA, beta-D-galactopyranose | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LND
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LN4
 
 | | Crystal structure of self-complemented PsaA, the major subunit of pH 6 antigen from Yersinia pests, in complex with choline | | Descriptor: | CHOLINE ION, pH 6 antigen,pH 6 antigen | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LO7
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
4PHX
 
 | |
