6HH0
 
 | |
1DFS
 
 | | SOLUTION STRUCTURE OF THE ALPHA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
1DFT
 
 | | SOLUTION STRUCTURE OF THE BETA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
2AP8
 
 | |
2AP7
 
 | |
2JTW
 
 | |
2KMT
 
 | |
2KV5
 
 | |
8OZZ
 
 | |
8ALO
 
 | | Heterodimer formation of sensory domains of Vibrio cholerae regulators ToxR and ToxS | | Descriptor: | Cholera toxin transcriptional activator, Transmembrane regulatory protein ToxS | | Authors: | Gubensaek, N, Sagmeister, T, Pavkov-Keller, T, Zangger, K, Buhlheller, C, Wagner, G.E. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Vibrio cholerae's ToxRS bile sensing system.
Elife, 12, 2023
|
|
2ADL
 
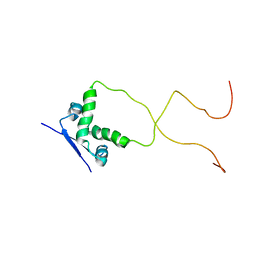 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADN
 
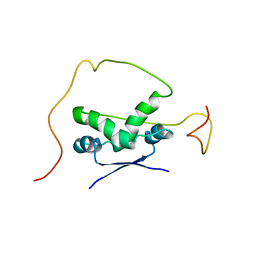 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
1JI9
 
 | |
1T2Y
 
 | | NMR solution structure of the protein part of Cu6-Neurospora crassa MT | | Descriptor: | Metallothionein | | Authors: | Cobine, P.A, McKay, R.T, Zangger, K, Dameron, C.T, Armitage, I.M. | | Deposit date: | 2004-04-23 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Cu metallothionein from the fungus Neurospora crassa
Eur.J.Biochem., 271, 2004
|
|
4L0J
 
 | | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems | | Descriptor: | DNA helicase I, MAGNESIUM ION, SULFATE ION | | Authors: | Redzej, A, Ilangovan, A, Lang, S, Gruber, C.J, Topf, M, Zangger, K, Zechner, E.L, Waksman, G. | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems.
Mol.Microbiol., 89, 2013
|
|
4CF6
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with Cibacron blue at 2.7 A resolution | | Descriptor: | CIBACRON BLUE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
2AN7
 
 | | Solution structure of the bacterial antidote ParD | | Descriptor: | Protein parD | | Authors: | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
4CET
 
 | | Crystal structure of the complex of the P187S variant of human NAD(P) H:quinone oxidoreductase with dicoumarol at 2.2 A resolution | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Gudipati, V, Uhl, M.K, Binter, A, Pulido, S, Saf, R, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Collapse of the Native Structure by a Single Amino Acid Exchange in Human Nad(P)H:Quinone Oxidoreductase (Nqo1).
FEBS J., 281, 2014
|
|
7NN6
 
 | |
7NMB
 
 | |
5A4K
 
 | | Crystal structure of the R139W variant of human NAD(P)H:quinone oxidoreductase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Strandback, E, Gudipati, V, Uhl, M.K, Rantase, D.M, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Catalytic competence, structure and stability of the cancer-associated R139W variant of the human NAD(P)H:quinone oxidoreductase 1 (NQO1).
FEBS J., 284, 2017
|
|
5AIW
 
 | |
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2KLG
 
 | | PERE NMR structure of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
