9GOJ
 
 | |
8AIR
 
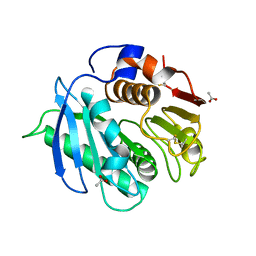 | | Crystal structure of cutinase RgCutII from Rhizobacter gummiphilus | | Descriptor: | ACETATE ION, RgCutII | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
6EUS
 
 | |
8AYV
 
 | |
8C65
 
 | |
4R3U
 
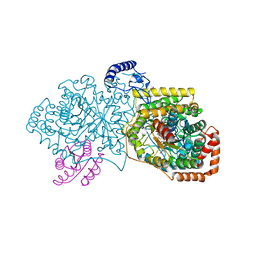 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
4RLC
 
 | |
4DGJ
 
 | |
4RLB
 
 | |
4RL9
 
 | |
3QNJ
 
 | |
9FF9
 
 | |
8R2V
 
 | | Crystal structure of hydroxyquinol-1,2-dioxygenase from Gelatoporia subvermispora (GsHDX1) | | Descriptor: | FE (III) ION, Intradiol ring-cleavage dioxygenases domain-containing protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Zahn, M, Kuatsjah, E, Salvachua, D. | | Deposit date: | 2023-11-07 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and structural characterization of enzymes in the 4-hydroxybenzoate catabolic pathway of lignin-degrading white-rot fungi.
Cell Rep, 43, 2024
|
|
8R2T
 
 | |
8R2W
 
 | | Crystal structure of hydroxyquinol-1,2-dioxygenase from Trametes versicolor (TvHDX1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, Hydroxyquinol 1,2-dioxygenase, ... | | Authors: | Zahn, M, Kuatsjah, E, Salvachua, D. | | Deposit date: | 2023-11-07 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Biochemical and structural characterization of enzymes in the 4-hydroxybenzoate catabolic pathway of lignin-degrading white-rot fungi.
Cell Rep, 43, 2024
|
|
8R2U
 
 | |
8R2X
 
 | | Crystal structure of hydroxyquinol-1,2-dioxygenase from Rhodococcus jostii RHA1 (RjTsdC) | | Descriptor: | 6-chlorohydroxyquinol-1,2-dioxygenase, FE (III) ION, PHOSPHATIDYLETHANOLAMINE | | Authors: | Zahn, M, Kuatsjah, E, Salvachua, D. | | Deposit date: | 2023-11-07 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Biochemical and structural characterization of enzymes in the 4-hydroxybenzoate catabolic pathway of lignin-degrading white-rot fungi.
Cell Rep, 43, 2024
|
|
5MDP
 
 | |
5MDS
 
 | |
4JWE
 
 | |
4JWD
 
 | |
5MDR
 
 | | Crystal structure of in vitro folded Chitoporin VhChip from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitoporin, ... | | Authors: | Zahn, M, van den Berg, B. | | Deposit date: | 2016-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for chitin acquisition by marine Vibrio species.
Nat Commun, 9, 2018
|
|
5MDO
 
 | |
4JWC
 
 | |
4JWI
 
 | |
