6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
1II9
 
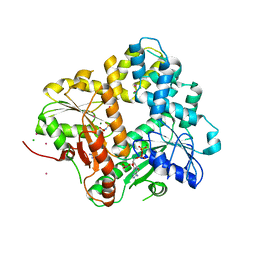 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH AMP-PNP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, CADMIUM ION, ... | | 著者 | Zhou, T, Radaev, S, Gatti, D.L, Rosen, B.P. | | 登録日 | 2001-04-21 | | 公開日 | 2001-09-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1FWL
 
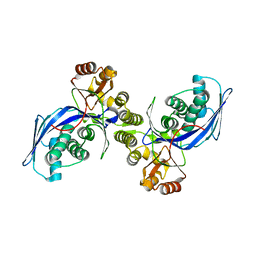 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE | | 分子名称: | HOMOSERINE KINASE | | 著者 | Zhou, T, Daugherty, M, Grishin, N.V, Osterman, A.L, Zhang, H. | | 登録日 | 2000-09-22 | | 公開日 | 2000-12-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily.
Structure Fold.Des., 8, 2000
|
|
1FWK
 
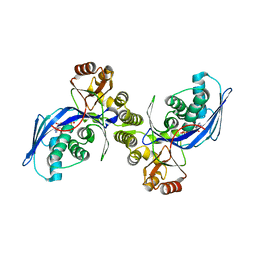 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HOMOSERINE KINASE, MAGNESIUM ION | | 著者 | Zhou, T, Daugherty, M, Grishin, N.V, Osterman, A.L, Zhang, H. | | 登録日 | 2000-09-22 | | 公開日 | 2000-12-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily.
Structure Fold.Des., 8, 2000
|
|
1IHU
 
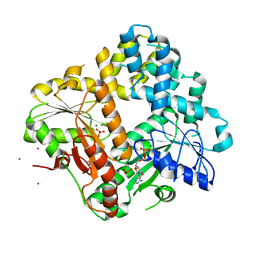 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH MG-ADP-ALF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ARSENICAL PUMP-DRIVING ATPASE, ... | | 著者 | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | 登録日 | 2001-04-20 | | 公開日 | 2001-09-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
2Z60
 
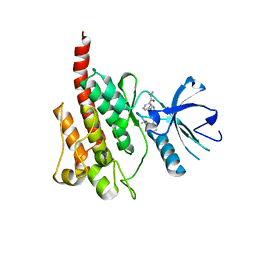 | | Crystal Structure of the T315I Mutant of Abl kinase bound with PPY-A | | 分子名称: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | 著者 | Zhou, T, Dalgarno, D, Zhu, X. | | 登録日 | 2007-07-21 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
7R74
 
 | |
7R73
 
 | |
7RI1
 
 | |
7RI2
 
 | |
4RX4
 
 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC134 in complex with HIV-1 clade A Q842.d12 gp120 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC134 Heavy chain, ... | | 著者 | Zhou, T, Acharya, P, Moquin, S, Kwong, P.D. | | 登録日 | 2014-12-08 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
8FBW
 
 | | Crystal structure of SIV-1 V2 antibody NCI05 in complex with a V2 peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Zhou, T, Kwong, P.D, Van Wazer, D.J. | | 登録日 | 2022-11-30 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Effect of Passive Administration of Monoclonal Antibodies Recognizing Simian Immunodeficiency Virus (SIV) V2 in CH59-Like Coil/Helical or beta-Sheet Conformations on Time of SIV mac251 Acquisition.
J.Virol., 97, 2023
|
|
7TCC
 
 | |
7LRT
 
 | |
7LRS
 
 | |
7U0D
 
 | |
7MLZ
 
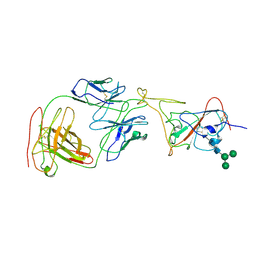 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | 分子名称: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | 著者 | Zhou, T, Tsybovsky, T, Kwong, P.D. | | 登録日 | 2021-04-29 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MM0
 
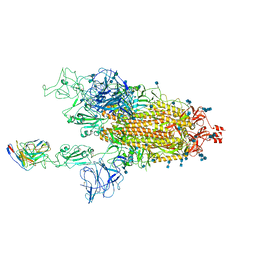 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | 著者 | Zhou, T, Tsybovsky, T, Kwong, P.D. | | 登録日 | 2021-04-29 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
5F96
 
 | |
7TCQ
 
 | |
6URM
 
 | | Crystal structure of vaccine-elicited receptor-binding site targeting antibody LPAF-a.01 in complex with Hemagglutinin H1 A/California/04/2009 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zhou, T, Cheung, S.F, Kwong, P.D. | | 登録日 | 2019-10-23 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation.
Cell Rep, 32, 2020
|
|
