1HMT
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, STEARIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HMS
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HOC
 
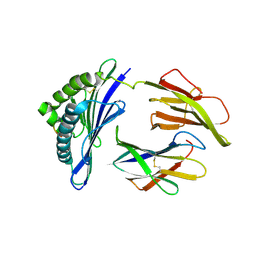 | | THE THREE-DIMENSIONAL STRUCTURE OF H-2DB AT 2.4 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ANTIGEN-DETERMINANT SELECTION | | Descriptor: | 9-RESIDUE PEPTIDE, BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H2-DB) (ALPHA CHAIN) | | Authors: | Young, A.C.M, Zhang, W, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection
Cell(Cambridge,Mass.), 76, 1994
|
|
1HMR
 
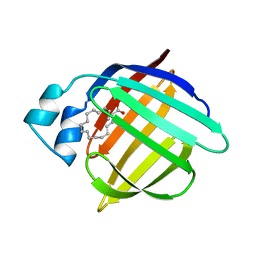 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | 9-OCTADECENOIC ACID, MUSCLE FATTY ACID BINDING PROTEIN | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
4L5E
 
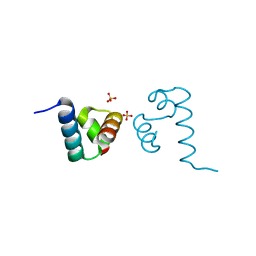 | | Crystal structure of A. aeolicus NtrC1 DNA binding domain | | Descriptor: | SULFATE ION, Transcriptional regulator (NtrC family) | | Authors: | Young, A, Maris, A.E, Vidangos, N.K, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
2H1P
 
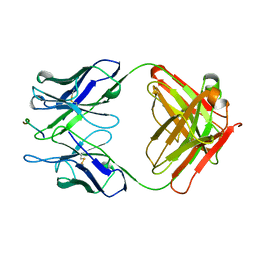 | | THE THREE-DIMENSIONAL STRUCTURES OF A POLYSACCHARIDE BINDING ANTIBODY TO CRYPTOCOCCUS NEOFORMANS AND ITS COMPLEX WITH A PEPTIDE FROM A PHAGE DISPLAY LIBRARY: IMPLICATIONS FOR THE IDENTIFICATION OF PEPTIDE MIMOTOPES | | Descriptor: | 2H1, PA1 | | Authors: | Young, A.C.M, Valadon, P, Casadevall, A, Scharff, M.D, Sacchettini, J.C. | | Deposit date: | 1997-11-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes.
J.Mol.Biol., 274, 1997
|
|
4O2X
 
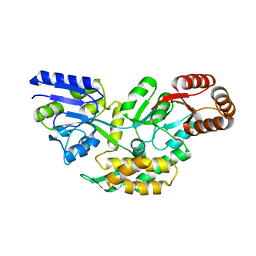 | | Structure of a malarial protein | | Descriptor: | Maltose-binding periplasmic protein, ATP-dependent Clp protease adaptor protein ClpS containing protein chimeric construct | | Authors: | AhYoung, A.P, Koehl, A, Cascio, D, Egea, P.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a putative ClpS N-end rule adaptor protein from the malaria pathogen Plasmodium falciparum.
Protein Sci., 25, 2016
|
|
4LYT
 
 | |
3LYT
 
 | |
6LYT
 
 | |
1LD9
 
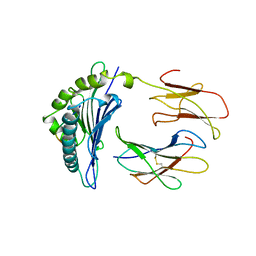 | | THE THREE-DIMENSIONAL STRUCTURE OF AN H-2LD PEPTIDE COMPLEX EXPLAINS THE UNIQUE INTERACTION OF LD WITH BETA2M AND PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2LD HEAVY CHAIN, NANO-PEPTIDE | | Authors: | Balendiran, G.K, Solheim, J.C, Young, A.C.M, Hansen, T.H, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1997-04-24 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of an H-2Ld-peptide complex explains the unique interaction of Ld with beta-2 microglobulin and peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5LYT
 
 | |
2MHA
 
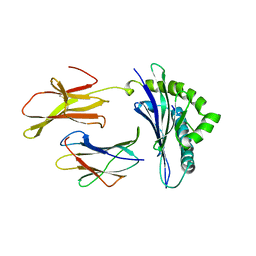 | | CRYSTAL STRUCTURE OF THE MAJOR HISTOCOMPATIBILITY COMPLEX CLASS I H-2KB MOLECULE CONTAINING A SINGLE VIRAL PEPTIDE: IMPLICATIONS FOR PEPTIDE BINDING AND T-CELL RECEPTOR RECOGNITION | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H-2KB) (ALPHA CHAIN), VIRAL OCTAPEPTIDE ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU | | Authors: | Zhang, W, Young, A.C.M, Imarai, M, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1993-07-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
8FCI
 
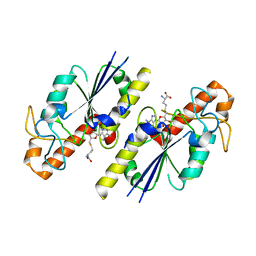 | | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lavallee, T, Mehta, K, Young, A, Cortes, J, Isaacson, B, Brylewski, J, Schlegel, F, Doti, L, Battaile, K, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine
To Be Published
|
|
8FCH
 
 | | Apo Structure of (N1G37) Methyltransferase from Mycobacterium avium | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Isaacson, B, Brylewski, J, Schlegel, F, Cortes, J, Lavallee, T, Mehta, K, Young, A, Doti, L, Battaile, K.P, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
To Be Published
|
|
2WVW
 
 | | Cryo-EM structure of the RbcL-RbcX complex | | Descriptor: | RBCX PROTEIN, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN | | Authors: | Liu, C, Young, A.L, Starling-Windhof, A, Bracher, A, Saschenbrecker, S, Rao, B.V, Rao, K.V, Berninghausen, O, Mielke, T, Hartl, F.U, Beckmann, R, Hayer-Hartl, M. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Coupled Chaperone Action in Folding and Assembly of Hexadecameric Rubisco
Nature, 463, 2010
|
|
4NJJ
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Manganese(II) | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJG
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxypterin | | Descriptor: | 6-CARBOXYPTERIN, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4L4U
 
 | | Crystal structure of construct containing A. aeolicus NtrC1 receiver, central and DNA binding domains | | Descriptor: | Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Maris, A.E, Young, A, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-09 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
4NJK
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 7-carboxy-7-deazaguanine, and Mg2+ | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJI
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Mg2+ | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJH
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxy-5,6,7,8-tetrahydropterin | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4XBI
 
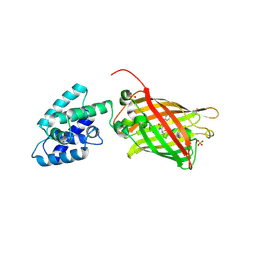 | | Structure Of A Malarial Protein Involved in Proteostasis | | Descriptor: | ClpB protein, putative,Green fluorescent protein, SULFATE ION | | Authors: | Egea, P.F, Ah Young, A.P, Cascio, D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural mapping of the ClpB ATPases of Plasmodium falciparum: Targeting protein folding and secretion for antimalarial drug design.
Protein Sci., 24, 2015
|
|
5VKZ
 
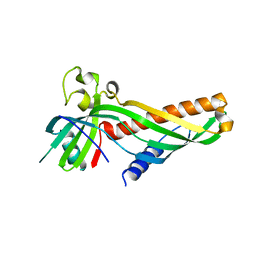 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
1XZ2
 
 | | wild-type hemoglobin deoxy no-salt | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A, Hui, H.L, Wierzba, A, DeYoung, A, Kwiatkowski, L.D, Noble, R.W, Juszczak, L.J, Peterson, E.S, Friedman, J.M. | | Deposit date: | 2004-11-11 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intersubunit interactions associated with tyr42alpha stabilize the quaternary-T tetramer but are not major quaternary constraints in deoxyhemoglobin
Biochemistry, 44, 2005
|
|
