3LQM
 
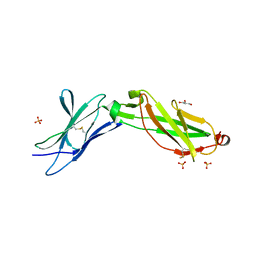 | | Structure of the IL-10R2 Common Chain | | Descriptor: | GLYCEROL, Interleukin-10 receptor subunit beta, SULFATE ION | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2010-02-09 | | Release date: | 2010-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and mechanism of receptor sharing by the IL-10R2 common chain.
Structure, 18, 2010
|
|
3MTX
 
 | | Crystal structure of chicken MD-1 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, GLYCEROL, Protein MD-1, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MU3
 
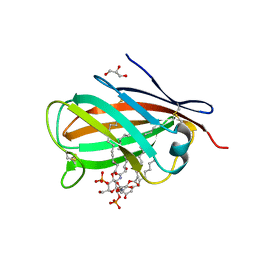 | | Crystal structure of chicken MD-1 complexed with lipid IVa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1Y6N
 
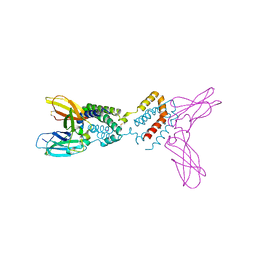 | | Crystal structure of Epstein-Barr virus IL-10 mutant (A87I) complexed with the soluble IL-10R1 chain | | Descriptor: | Interleukin-10 receptor alpha chain, Viral interleukin-10 homolog | | Authors: | Yoon, S.I, Jones, B.C, Logsdon, N.J, Walter, M.R. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain.
Structure, 13, 2005
|
|
1Y6M
 
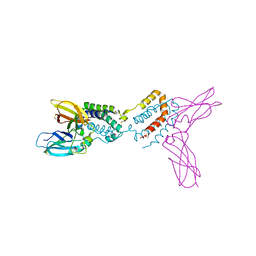 | | Crystal structure of Epstein-Barr virus IL-10 complexed with the soluble IL-10R1 chain | | Descriptor: | Interleukin-10 receptor alpha chain, Viral interleukin-10 homolog | | Authors: | Yoon, S.I, Jones, B.C, Logsdon, N.J, Walter, M.R. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain.
Structure, 13, 2005
|
|
1Y6K
 
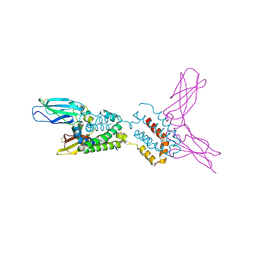 | | Crystal structure of human IL-10 complexed with the soluble IL-10R1 chain | | Descriptor: | Interleukin-10, Interleukin-10 receptor alpha chain | | Authors: | Yoon, S.I, Jones, B.C, Josepson, K, Logsdon, N.J, Walter, M.R. | | Deposit date: | 2004-12-06 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain.
Structure, 13, 2005
|
|
3RG1
 
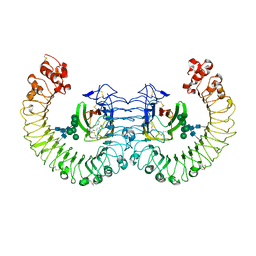 | | Crystal structure of the RP105/MD-1 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CD180 molecule, LY86 protein, ... | | Authors: | Yoon, S.I, Hong, M, Wilson, I.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2H24
 
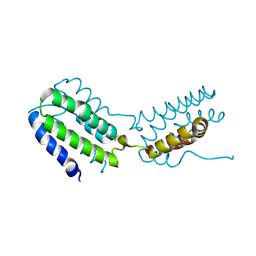 | | Crystal structure of human IL-10 | | Descriptor: | Interleukin-10 | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2006-05-18 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex.
J.Biol.Chem., 281, 2006
|
|
3V47
 
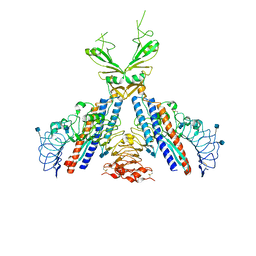 | | Crystal structure of the N-terminal fragment of zebrafish TLR5 in complex with Salmonella flagellin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Flagellin, ... | | Authors: | Yoon, S.I, Hong, H, Wilson, I.A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis of TLR5-flagellin recognition and signaling.
Science, 335, 2012
|
|
3V44
 
 | |
6J3D
 
 | |
4XPK
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH | | Descriptor: | N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
4XPL
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
8J56
 
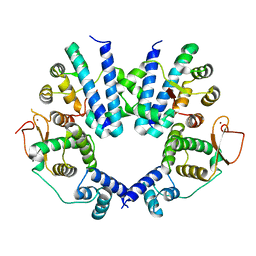 | | Crystal structure of the FlhDC complex from Cupriavidus necator | | Descriptor: | Flagellar transcriptional regulator FlhC, Flagellar transcriptional regulator FlhD, ZINC ION | | Authors: | Cho, S.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-04-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
8K3F
 
 | |
8JZC
 
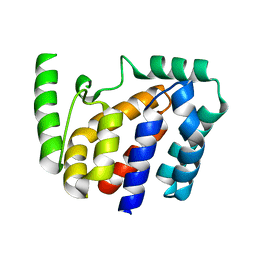 | | Crystal structure of Geobacillus stearothermophilus NarJ | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
8JZD
 
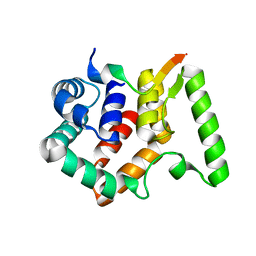 | | Crystal structure of Escherichia coli NarJ in complex with the signal peptide of E. coli NarG | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone NarJ, Respiratory nitrate reductase 1 alpha chain | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
7F2H
 
 | |
7F2G
 
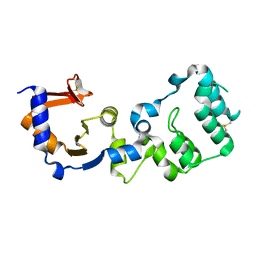 | |
8GR2
 
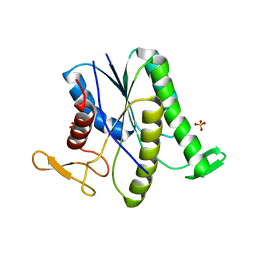 | |
8H50
 
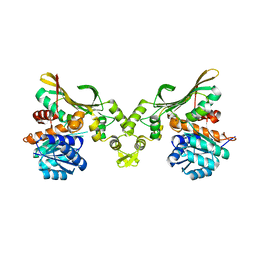 | |
8H4Z
 
 | |
8H52
 
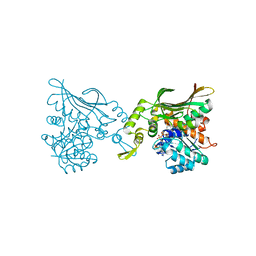 | | Crystal structure of Helicobacter pylori carboxyspermidine dehydrogenase in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | Authors: | Ko, K.Y, Park, S.C, Cho, S.Y, Yoon, S.I. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of carboxyspermidine dehydrogenase from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
8IEU
 
 | |
8IEV
 
 | |
