1DN9
 
 | |
1F42
 
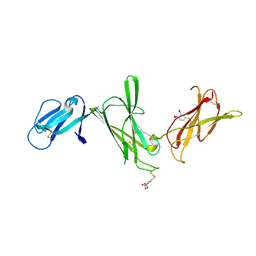 | | THE P40 DOMAIN OF HUMAN INTERLEUKIN-12 | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
1F45
 
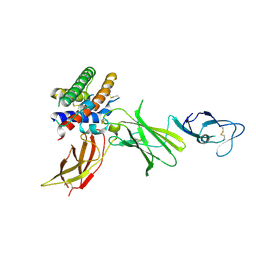 | | HUMAN INTERLEUKIN-12 | | Descriptor: | INTERLEUKIN-12 ALPHA CHAIN, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
6BNA
 
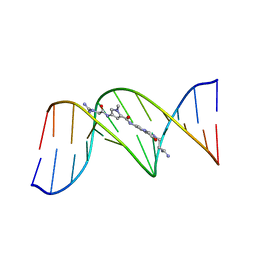 | | BINDING OF AN ANTITUMOR DRUG TO DNA. NETROPSIN AND C-G-C-G-A-A-T-T-BRC-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*CP*G)-3'), NETROPSIN | | Authors: | Kopka, M.L, Yoon, C, Goodsell, D, Pjura, P, Dickerson, R.E. | | Deposit date: | 1984-08-30 | | Release date: | 1984-10-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G.
J.Mol.Biol., 183, 1985
|
|
7X7H
 
 | | Crystal structure of Fructose regulator/Histidine phosphocarrier protein complex from Vibrio cholerae | | Descriptor: | CALCIUM ION, Catabolite repressor/activator, HPr family phosphocarrier protein | | Authors: | Kim, M.-K, Zhang, J, Yoon, C.-K, Seok, Y.-J. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HPr prevents FruR-mediated facilitation of RNA polymerase binding to the fru promoter in Vibrio cholerae.
Nucleic Acids Res., 51, 2023
|
|
6L2U
 
 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | Authors: | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | Deposit date: | 2019-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
7BER
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly (solved, rebuilt and refined using an identical protocol to the MicroED structure of the MyD88 TIR domain higher-order assembly) | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7BEQ
 
 | | MicroED structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
4RWA
 
 | | Synchrotron structure of the human delta opioid receptor in complex with a bifunctional peptide (PSI community target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Soluble cytochrome b562,Delta-type opioid receptor, bifunctional peptide | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RWD
 
 | | XFEL structure of the human delta opioid receptor in complex with a bifunctional peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, SODIUM ION, ... | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RVY
 
 | | Serial Time resolved crystallography of Photosystem II using a femtosecond X-ray laser. The S state after two flashes (S3) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.-H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-11-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
7LAV
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LAX
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LB9
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase (G150T-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
1ANA
 
 | |
7LDO
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-3) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LDI
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-2) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LDM
 
 | | G150T Pseudomonas fluorescens isocyanide hydratase (G150T-1) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9S
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9W
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA0
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-2) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LD6
 
 | | G150A Pseudomonas fluorescens isocyanide hydratase (G150A-1) at 274K, Phenix-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Q
 
 | | Wild-type Pseudomonas fluorescens isocyanide hydratase (WT-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-04 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7L9Z
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-1) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
7LA3
 
 | | Pseudomonas fluorescens G150A isocyanide hydratase (G150A-3) at 274K, Refmac5-refined | | Descriptor: | Isonitrile hydratase InhA | | Authors: | Su, Z, Dasgupta, M, Poitevin, F, Mathews, I.I, van den Bedem, H, Wall, M.E, Yoon, C.H, Wilson, M.A. | | Deposit date: | 2021-01-05 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Reproducibility of protein x-ray diffuse scattering and potential utility for modeling atomic displacement parameters.
Struct Dyn., 8, 2021
|
|
