2FH8
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with isomaltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHC
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotriose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FH6
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with glucose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2QHT
 
 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2FGZ
 
 | | Crystal Structure Analysis of apo pullulanase from Klebsiella pneumoniae | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
7VOV
 
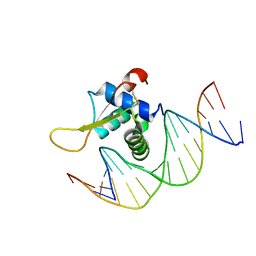
 | | The crystal structure of human forkhead box protein in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOU
 
 | | The crystal structure of human forkhead box protein in complex with DNA 1 | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VHV
 
 | | Crystal structure of S. aureus D-alanine alanyl carrier protein ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanyl carrier protein ligase, MAGNESIUM ION | | Authors: | Lee, B.J, Lee, I.-G, Im, H.G, Yoon, H.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of the D-alanyl carrier protein ligase DltA from Staphylococcus aureus Mu50.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1LXL
 
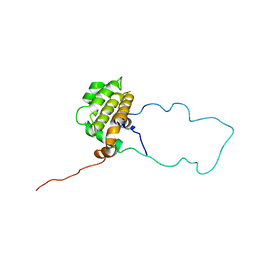 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1PX8
 
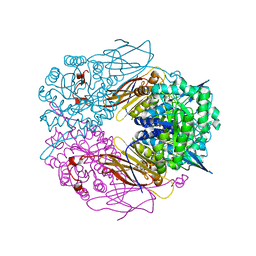 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
2QHS
 
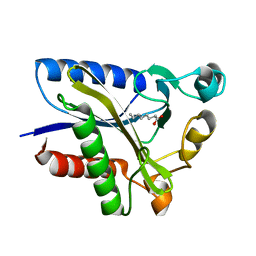 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2QHV
 
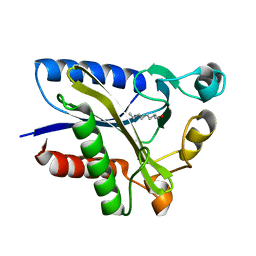 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTAN-1-OL | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
1WOG
 
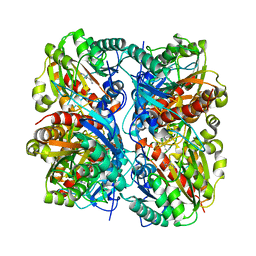 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOI
 
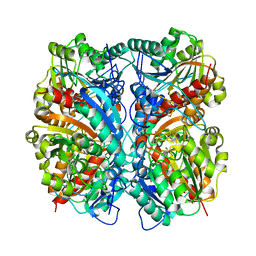 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOH
 
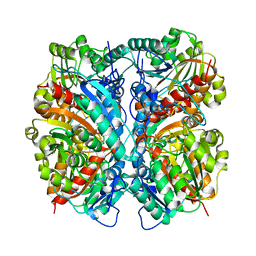 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
5GNP
 
 | |
1DHM
 
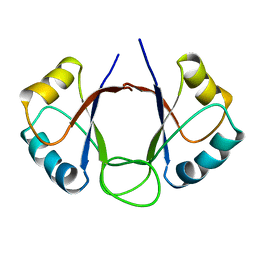 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
3CNO
 
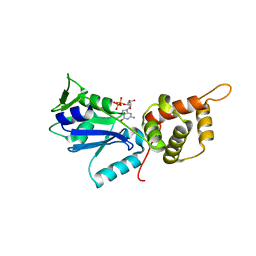 | | GDP-bound structue of TM YlqF | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
1UAJ
 
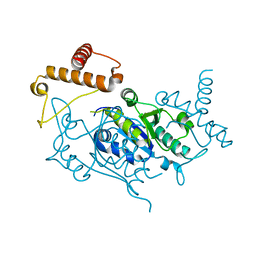 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAM
 
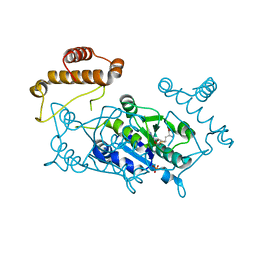 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
3CNN
 
 | | GTP-bound structure of TM YlqF | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3QY8
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
2O4C
 
 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
3CNL
 
 | | Crystal structure of GNP-bound YlqF from T. maritima | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | Deposit date: | 2009-07-31 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
