5G12
 
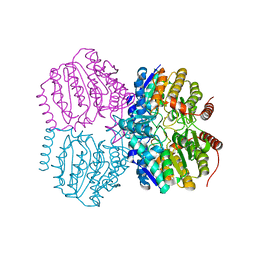 | |
4CBK
 
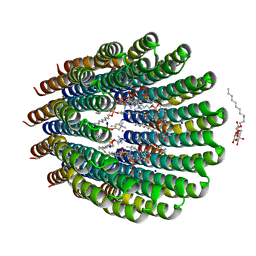 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4CDB
 
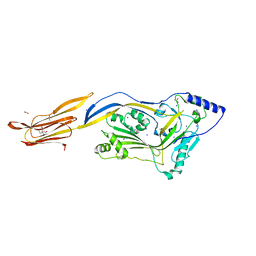 | | Crystal structure of listeriolysin O | | Descriptor: | ACETATE ION, LISTERIOLYSIN O, SODIUM ION, ... | | Authors: | Koester, S, Yildiz, O. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Listeriolysin O Reveals Molecular Details of Oligomerization and Pore Formation
Nat.Commun., 5, 2014
|
|
4CBJ
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4BEM
 
 | | Crystal structure of the F-type ATP synthase c-ring from Acetobacterium woodii. | | Descriptor: | ACETATE ION, F1FO ATPASE C1 SUBUNIT, F1FO ATPASE C2 SUBUNIT, ... | | Authors: | Matthies, D, Meier, T, Yildiz, O. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Structure and Mechanism of an F/V-Hybrid Rotor Ring in a Na+-Coupled ATP Synthase
Nat.Commun., 5, 2014
|
|
4CZA
 
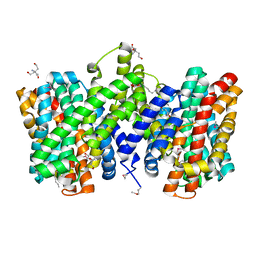 | | Structure of the sodium proton antiporter PaNhaP from Pyrococcus abyssii with bound thallium ion. | | Descriptor: | ACETATE ION, NA+/H+ ANTIPORTER, PUTATIVE, ... | | Authors: | Woehlert, D, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and substrate ion binding in the sodium/proton antiporter PaNhaP.
Elife, 3, 2014
|
|
4CZ8
 
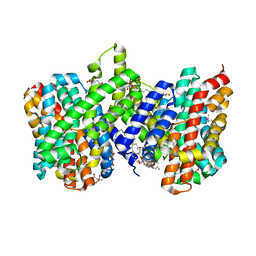 | | Structure of the sodium proton antiporter PaNhaP from Pyrococcus abyssii at pH 8. | | Descriptor: | CITRATE ANION, NA+/H+ ANTIPORTER, PUTATIVE, ... | | Authors: | Woehlert, D, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure and Substrate Ion Binding in the Sodium/Proton Antiporter Panhap.
Elife, 3, 2014
|
|
4D0A
 
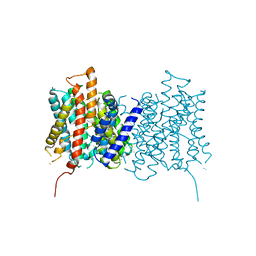 | |
4AIN
 
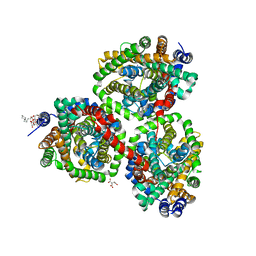 | | Crystal structure of BetP with asymmetric protomers. | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Koshy, C, Ziegler, C, Yildiz, O. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Alternating-Access Mechanism in Conformationally Asymmetric Trimers of the Betaine Transporter Betp.
Nature, 490, 2012
|
|
4CZB
 
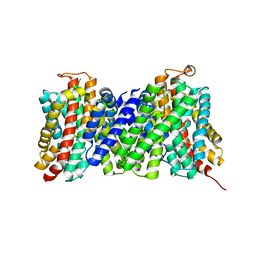 | | Structure of the sodium proton antiporter MjNhaP1 from Methanocaldococcus jannaschii at pH 8. | | Descriptor: | NA(+)/H(+) ANTIPORTER 1, POTASSIUM ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Woehlert, D, Paulino, C, Kapotova, E, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 3D Em Map of the Sodium Proton Antiporter Mjnhap1 from Methanocaldococcus Jannaschii
Elife, 3, 2014
|
|
4CZ9
 
 | |
7A73
 
 | |
7AQW
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (membrane tip) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Acyl carrier protein 1, mitochondrial, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AQQ
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AQR
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (peripheral arm) | | Descriptor: | Acyl carrier protein 2, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARC
 
 | | Cryo-EM structure of Polytomella Complex-I (peripheral arm) | | Descriptor: | 13 kDa, 18 kDa, 24 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR9
 
 | | Cryo-EM structure of Polytomella Complex-I (membrane arm) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 15 kDa, AGGG, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARD
 
 | | Cryo-EM structure of Polytomella Complex-I (complete composition) | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 13 kDa, 15 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARB
 
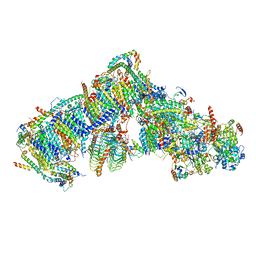 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7AR8
 
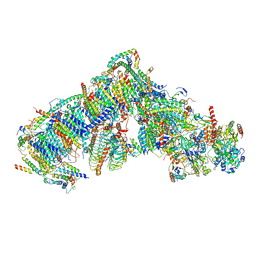 | | Cryo-EM structure of Arabidopsis thaliana complex-I (closed conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7B4L
 
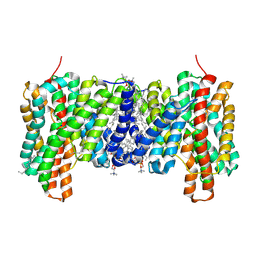 | |
7B4M
 
 | |
6RDK
 
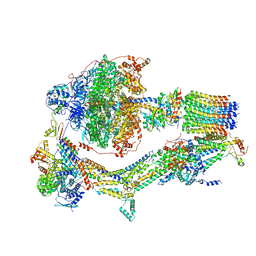 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1B, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE0
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REP
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 3, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
