6UG6
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDW
 
 | | S2 symmetric peptide design number 3 crystal form 2, Lurch | | Descriptor: | S2-3, Lurch crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGB
 
 | | C3 symmetric peptide design number 2, Baby Basil | | Descriptor: | C3 symmetric peptide design number 2, Baby Basil, CHLORIDE ION, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF8
 
 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFU
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG3
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UD9
 
 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UCX
 
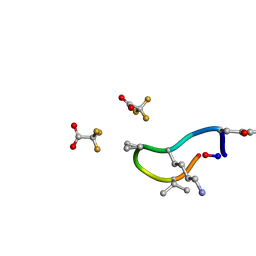 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
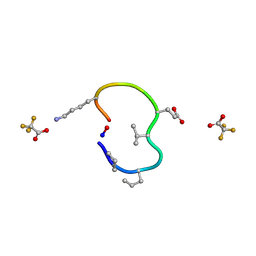 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
4QIF
 
 | | Crystal Structure of PduA with edge mutation K26A and pore mutation S40H | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Pang, A.H, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-05-30 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9951 Å) | | Cite: | Selective molecular transport through the protein shell of a bacterial microcompartment organelle.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QF0
 
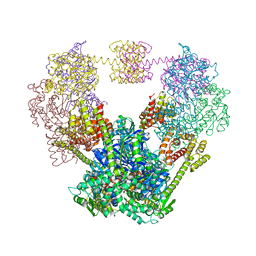 | |
4QES
 
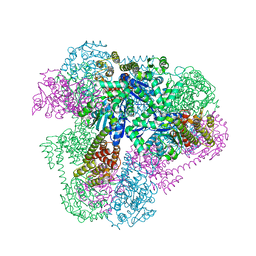 | |
4QCC
 
 | |
4QIG
 
 | |
4QFF
 
 | |
4QIV
 
 | |
4QIE
 
 | |
4RBT
 
 | |
4RBV
 
 | |
4RBU
 
 | |
4RC1
 
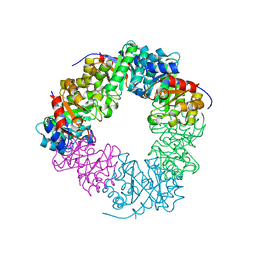 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii with PRPP | | Descriptor: | PHOSPHATE ION, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E.J, Cascio, D, Sawaya, M.R, Yeates, T.O, Rasche, M.E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4TME
 
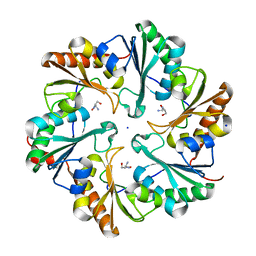 | |
