3A8E
 
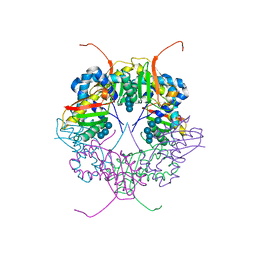 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1V54
 
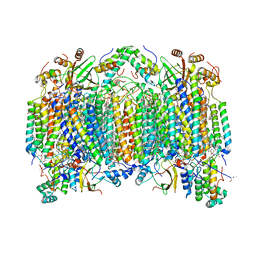 | | Bovine heart cytochrome c oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
1V55
 
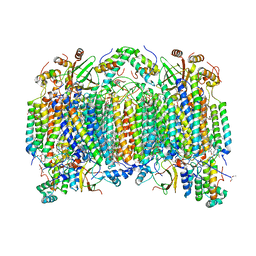 | | Bovine heart cytochrome c oxidase at the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1UCC
 
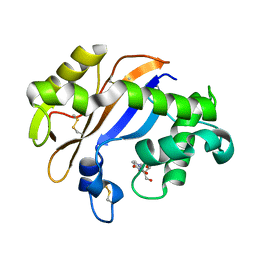 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
2D74
 
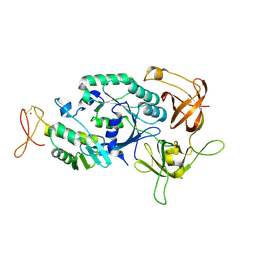 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer | | Descriptor: | Translation initiation factor 2 beta subunit, Translation initiation factor 2 gamma subunit, ZINC ION | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2005-11-16 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3AJ1
 
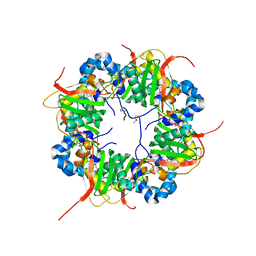 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
1V7O
 
 | |
1WMI
 
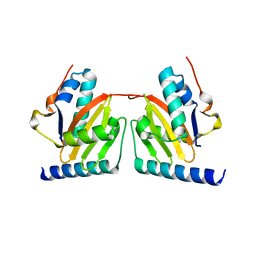 | | Crystal structure of archaeal RelE-RelB complex from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PHS013, hypothetical protein PHS014 | | Authors: | Takagi, H, Kakuta, Y, Kamachi, R, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WNU
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with L-serine | | Descriptor: | SERINE, ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X0T
 
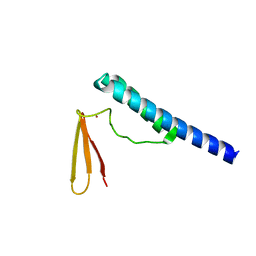 | | Crystal structure of ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3 | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Kakuta, Y, Ishimatsu, I, Numata, T, Kimura, K, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2005-03-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21(,)
Biochemistry, 44, 2005
|
|
5X32
 
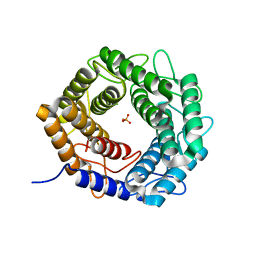 | | Crystal structure of D-mannose isomerase | | Descriptor: | N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Biochemical and structural characterization of Marinomonas mediterranead-mannose isomerase Marme_2490 phylogenetically distant from known enzymes
Biochimie, 144, 2018
|
|
5XW7
 
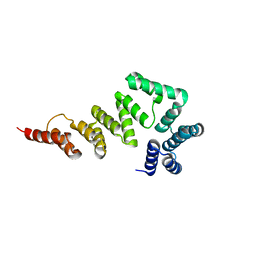 | |
5ZCC
 
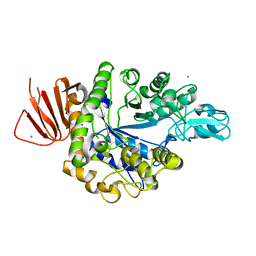 | | Crystal structure of Alpha-glucosidase in complex with maltose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5ZCE
 
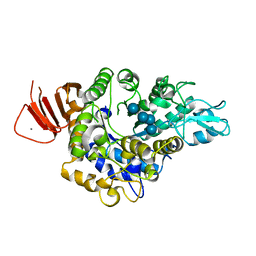 | | Crystal structure of Alpha-glucosidase in complex with maltotetraose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5ZCD
 
 | | Crystal structure of Alpha-glucosidase in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
2DI3
 
 | |
1UMJ
 
 | | Crystal structure of Pyrococcus horikoshii CutA in the presence of 3M guanidine hydrochloride | | Descriptor: | GUANIDINE, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Sakai, N, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2003-10-02 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for guanidine-protein side chain interactions: crystal structure of CutA from Pyrococcus horikoshii in 3M guanidine hydrochloride
Biochem.Biophys.Res.Commun., 323, 2004
|
|
3RXO
 
 | | Crystal structure of Trypsin complexed with (3-pyrrol-1-ylphenyl)methanamine | | Descriptor: | 1-[3-(1H-pyrrol-1-yl)phenyl]methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXI
 
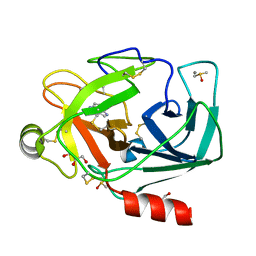 | | Crystal structure of Trypsin complexed with 2-(1H-indol-3-yl)ethanamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXL
 
 | | Crystal structure of Trypsin complexed with (2,5-dimethyl-3-furyl)methanamine | | Descriptor: | 1-(2,5-dimethylfuran-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXA
 
 | | Crystal structure of Trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXK
 
 | | Crystal structure of Trypsin complexed with methyl 4-amino-1-methyl-pyrrolidine-2-carboxylate | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXV
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and F03, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
