2DI3
 
 | |
1VE0
 
 | | Crystal structure of uncharacterized protein ST2072 from Sulfolobus tokodaii | | Descriptor: | SULFATE ION, ZINC ION, hypothetical protein (ST2072) | | Authors: | Tanabe, E, Horiike, Y, Tsumoto, K, Yasutake, Y, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the uncharacterized protein ST2072 from Sulfolobus tokodaii
To be Published
|
|
2D6Y
 
 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F85
 
 | | Structure analysis of Geranyl diphosphate methyltransferase | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2DGD
 
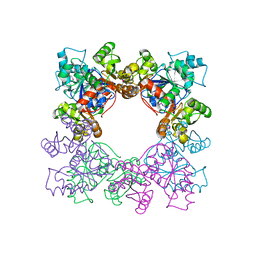 | | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii | | Descriptor: | 223aa long hypothetical arylmalonate decarboxylase, GLYCEROL | | Authors: | Tanaka, Y, Sasaki, T, Tanabe, E, Yao, M, Tanaka, I, Kumagai, I, Tsumoto, K. | | Deposit date: | 2006-03-10 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii
To be Published
|
|
2DF4
 
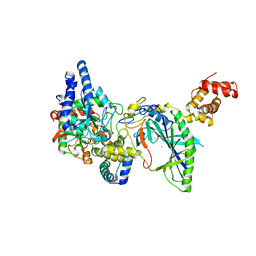 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Mn2+ | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
2DG7
 
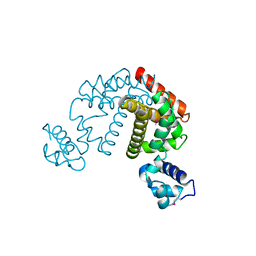 | | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2)
To be Published
|
|
2DG8
 
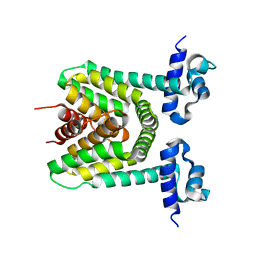 | | Crystal structure of the putative trasncriptional regulator SCO7518 from Streptomyces coelicolor A3(2) | | Descriptor: | putative tetR-family transcriptional regulatory protein | | Authors: | Hayashi, T, Watanabe, N, Sakai, N, Tamura, T, Yao, M, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO7518 from Streptomyces coelicolor A3(2)
To be Published
|
|
2DQN
 
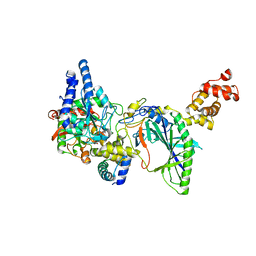 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Asn | | Descriptor: | ASPARAGINE, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2006-05-29 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
2DU9
 
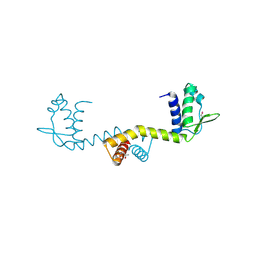 | | crystal structure of the transcriptional factor from C.glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Predicted transcriptional regulators | | Authors: | Gao, Y, Yao, M, Tanaka, I. | | Deposit date: | 2006-07-20 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structures of transcription factor CGL2947 from Corynebacterium glutamicum in two crystal forms: A novel homodimer assembling and the implication for effector-binding mode
Protein Sci., 16, 2007
|
|
4F84
 
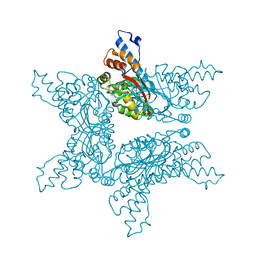 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with SAM | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2E8G
 
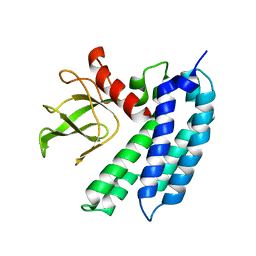 | |
2EK5
 
 | |
2F2A
 
 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Gln | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, GLUTAMINE, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
2DG6
 
 | | Crystal structure of the putative transcriptional regulator SCO5550 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and genomic DNA analysis of a putative transcription factor SCO5550 from Streptomyces coelicolor A3(2): regulating the expression of gene sco5551 as a transcriptional activator with a novel dimer shape
Biochem. Biophys. Res. Commun., 435, 2013
|
|
8IN4
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
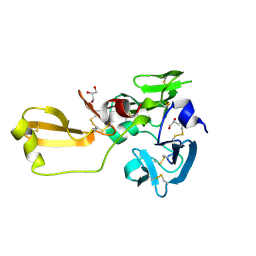 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
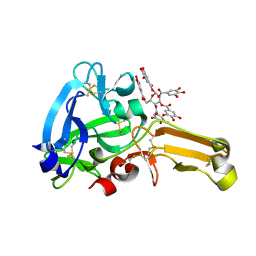 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
4I86
 
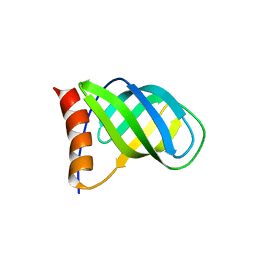 | | Crystal structure of PilZ domain of CeSA from cellulose synthesizing bacterium | | Descriptor: | Cellulose synthase 1 | | Authors: | Fujiwara, T, Komoda, K, Sakurai, N, Tanaka, I, Yao, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A
Biochem.Biophys.Res.Commun., 431, 2013
|
|
8H1K
 
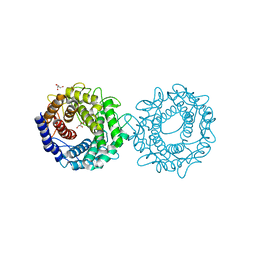 | | Crystal structure of glucose-2-epimerase from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, GLYCEROL, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1L
 
 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1M
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8IN1
 
 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
