3CU8
 
 | | Impaired binding of 14-3-3 to Raf1 is linked to Noonan and LEOPARD syndrome | | Descriptor: | 14-3-3 protein zeta/delta, MAGNESIUM ION, PROPANOIC ACID, ... | | Authors: | Schumacher, B, Weyand, M, Kuhlmann, J, Ottmann, C. | | Deposit date: | 2008-04-16 | | Release date: | 2009-05-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol. Cell. Biol., 30, 2010
|
|
4Z9W
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
1Y8I
 
 | | Horse methemoglobin low salt, PH 7.0 (98% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
4ZGR
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with T-Antigen. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
5B6F
 
 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
1Y8K
 
 | | Horse methemoglobin low salt, PH 7.0 (88% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8H
 
 | | HORSE METHEMOGLOBIN LOW SALT, PH 7.0 | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
3GNU
 
 | | Toxin fold as basis for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, CHLORIDE ION, GUANIDINE | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GNZ
 
 | | Toxin fold for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3P8D
 
 | | Crystal structure of the second Tudor domain of human PHF20 (homodimer form) | | Descriptor: | Medulloblastoma antigen MU-MB-50.72 | | Authors: | Cui, G, Lee, J, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2010-10-13 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3IQU
 
 | |
3IQJ
 
 | | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (10mer) | | Descriptor: | 10-mer peptide from RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Ottmann, C, Weyand, M. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol.Cell.Biol., 30, 2010
|
|
3DPU
 
 | | RocCOR domain tandem of Rab family protein (Roco) | | Descriptor: | Rab family protein | | Authors: | Gotthardt, K, Weyand, M, Kortholt, A, Van Haastert, P.J.M, Wittinghofer, A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase
Embo J., 27, 2008
|
|
3DPT
 
 | | COR domain of Rab family protein (Roco) | | Descriptor: | Rab family protein | | Authors: | Gotthardt, K, Weyand, M, Kortholt, A, Van Haastert, P.J.M, Wittinghofer, A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase
Embo J., 27, 2008
|
|
3PCV
 
 | | Crystal structure analysis of human leukotriene C4 synthase at 1.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The catalytic architecture of leukotriene C4 synthase with two arginine residues
J.Biol.Chem., 286, 2011
|
|
5VX7
 
 | |
1BWU
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | PROTEIN (AGGLUTININ), alpha-D-mannopyranose | | Authors: | Chandra, N.R, Ramachandraiah, G, Bachhawat, K, Dam, T.K, Surolia, A, Vijayan, M. | | Deposit date: | 1998-09-28 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a dimeric mannose-specific agglutinin from garlic: quaternary association and carbohydrate specificity.
J.Mol.Biol., 285, 1999
|
|
5TBN
 
 | |
1BZW
 
 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
5UMS
 
 | |
9EPR
 
 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Gi heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
3IQV
 
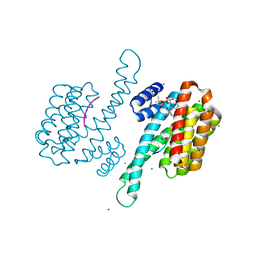 | |
5UMT
 
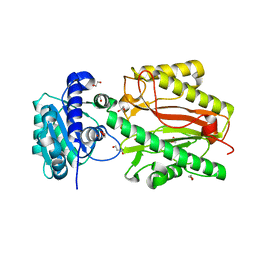 | | Crystal structure of N-terminal domain of human FACT complex subunit SPT16 | | Descriptor: | 1,2-ETHANEDIOL, FACT complex subunit SPT16, GLYCEROL | | Authors: | Su, D, Hu, Q, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SPT16
To Be Published
|
|
5UMV
 
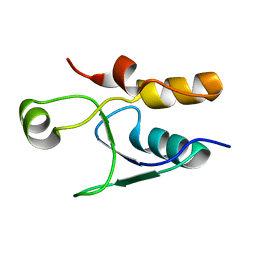 | |
5VZM
 
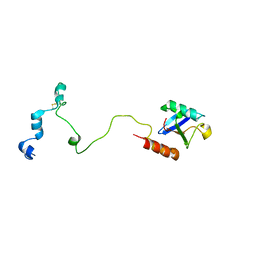 | |
