2H2Z
 
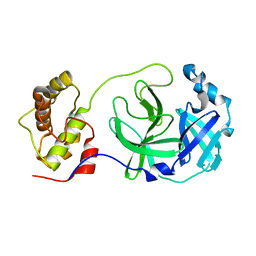 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
7D2T
 
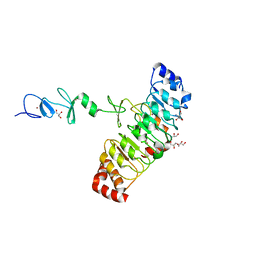 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Yang, H, Wei, Z, Yu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2S
 
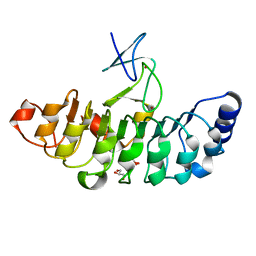 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
6N4F
 
 | |
6N4D
 
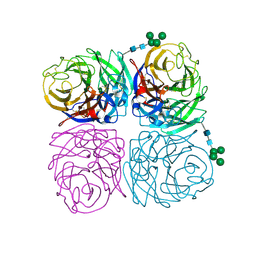 | | The crystal structure of neuramindase from A/canine/IL/11613/2015 (H3N2) influenza virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Assessment of Molecular, Antigenic, and Pathological Features of Canine Influenza A(H3N2) Viruses That Emerged in the United States.
J. Infect. Dis., 216, 2017
|
|
6N6B
 
 | |
1UK4
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6P40
 
 | | Crystal Structure of Full Length APOBEC3G FKL | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
1UJ1
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK2
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 | | Descriptor: | 3C-LIKE PROTEINASE | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK3
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MIU
 
 | | Structure of a BRCA2-DSS1 complex | | Descriptor: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
6P3X
 
 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.0) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6P3Z
 
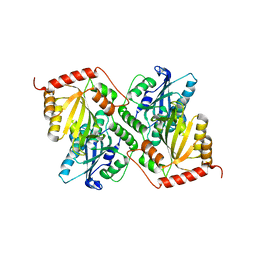 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 5.2) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6V48
 
 | | The crystal structure of hemagglutinin from A/mallard/Gurjev/263/1982 (H14N5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Molecular characterization and three-dimensional structures of avian H8, H11, H14, H15 and swine H4 influenza virus hemagglutinins
Heliyon, 6, 2020
|
|
6P3Y
 
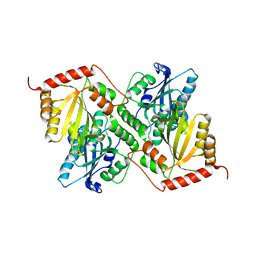 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
3NK6
 
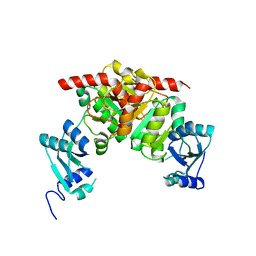 | | Structure of the Nosiheptide-resistance methyltransferase | | Descriptor: | 23S rRNA methyltransferase | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3NK7
 
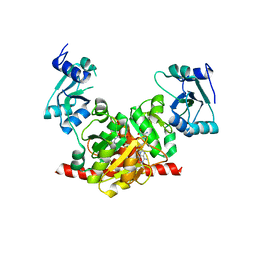 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | Descriptor: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
1D4N
 
 | | HUMAN SERUM TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, TRANSFERRIN | | Authors: | Yang, H.-W, MacGillivray, R.T.A, Chen, J, Luo, Y, Wang, Y, Brayer, G.D, Mason, A, Woodworth, R.C, Murphy, M.E.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-03-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two mutants (K206Q, H207E) of the N-lobe of human transferrin with increased affinity for iron.
Protein Sci., 9, 2000
|
|
1D3K
 
 | | HUMAN SERUM TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Yang, H.-W, MacGillivray, R.T.A, Chen, J, Luo, Y, Wang, Y, Brayer, G.D, Mason, A, Woodworth, R.C, Murphy, M.E.P. | | Deposit date: | 1999-09-29 | | Release date: | 2000-03-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two mutants (K206Q, H207E) of the N-lobe of human transferrin with increased affinity for iron.
Protein Sci., 9, 2000
|
|
1HY2
 
 | | MINIPROTEIN MP-1 COMPLEX WITH STREPTAVIDIN | | Descriptor: | MP-1, STREPTAVIDIN | | Authors: | Yang, H.W, Liu, D.Q, Fan, X, White, M.A, Fox, R.O. | | Deposit date: | 2001-01-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Ensemble Analysis of Ligand Binding in Streptavidin Mini-Protein Complexes
To be Published
|
|
4WSX
 
 | | The crystal structure of hemagglutinin from A/Jiangxi-Donghu/346/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
4WST
 
 | | The crystal structure of hemagglutinin from A/Taiwan/1/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
1HXL
 
 | | MINIPROTEIN MP-2 (V10A) COMPLEX WITH STREPTAVIDIN | | Descriptor: | MP-2, STREPTAVIDIN | | Authors: | Yang, H.W, Liu, D.Q, Fan, X, White, M.A, Fox, R.O. | | Deposit date: | 2001-01-16 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Ensemble Analysis of Ligand Binding in Streptavidin Mini-Protein Complexes
To be Published
|
|
