7VP9
 
 | | Crystal structure of human ClpP in complex with ZG111 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
5YOU
 
 | | Crystal structure of BRD4-BD1 bound with hjp64 | | Descriptor: | (3~{R})-4-cyclopropyl-~{N},1,3-trimethyl-~{N}-(4-methylphenyl)-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Bing, X, Jianping, H, Yanlian, L, Danyan, C. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Crystal structure of BRD4-BD1 bound with hjp64
To Be Published
|
|
4IWM
 
 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in P21 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
8HYN
 
 | | Bacterial STING from Riemerella anatipestifer | | Descriptor: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
6KKP
 
 | |
8YM4
 
 | |
8YM5
 
 | |
8YNI
 
 | |
8YNK
 
 | |
8YNL
 
 | |
8YNM
 
 | |
8YNN
 
 | |
2NOV
 
 | | Breakage-reunion domain of S.pneumoniae topo IV: crystal structure of a gram-positive quinolone target | | Descriptor: | DNA topoisomerase 4 subunit A | | Authors: | Laponogov, I, Veselkov, D.A, Sohi, M.K, Pan, X.S, Achari, A, Yang, C, Ferrara, J.D, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target.
PLoS ONE, 2, 2007
|
|
5ZLT
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Ko, T.P, Hsieh, T.J, Yang, C.S, Chen, Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
5YAW
 
 | |
3RZM
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*AP*TP*GP*TP*AP*TP*AP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZK
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(EDA)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
2JK1
 
 | | Crystal structure of the wild-type HupR receiver domain | | Descriptor: | HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
1YYE
 
 | | Crystal structure of estrogen receptor beta complexed with way-202196 | | Descriptor: | 3-(3-FLUORO-4-HYDROXYPHENYL)-7-HYDROXY-1-NAPHTHONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2005-02-24 | | Release date: | 2006-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
3RZJ
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(ME6)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
1YY4
 
 | | Crystal structure of estrogen receptor beta complexed with 1-chloro-6-(4-hydroxy-phenyl)-naphthalen-2-ol | | Descriptor: | 1-CHLORO-6-(4-HYDROXYPHENYL)-2-NAPHTHOL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2005-02-23 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
2JE2
 
 | | Cytochrome P460 from Nitrosomonas europaea - probable nonphysiological oxidized form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
2JE3
 
 | | Cytochrome P460 from Nitrosomonas europaea - probable physiological form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
2GBZ
 
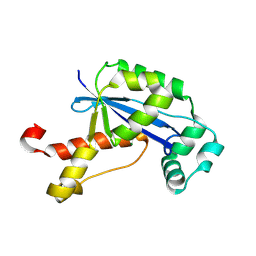 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
3ST9
 
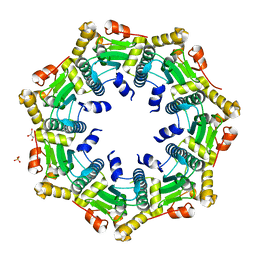 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
