6UCX
 
 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
3H8Y
 
 | |
3HLC
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, S5 mutant, unliganded | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLD
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant complex with monacolin J acid | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, FORMIC ACID, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLG
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with lovastatin | | Descriptor: | (3R,5R)-7-((1R,2R,6S,8R,8AS)-2,6-DIMETHYL-8-{[(2R)-2-METHYLBUTANOYL]OXY}-1,2,6,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL)-3,5-DIHYDROXYHEPTANOIC ACID, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLE
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with monacolin J acid | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLF
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with simvastatin | | Descriptor: | Simvastatin acid, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HL9
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, unliganded | | Descriptor: | Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLB
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, unliganded, selenomethionyl derivative | | Descriptor: | SULFATE ION, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3I6P
 
 | |
3IA0
 
 | |
3I96
 
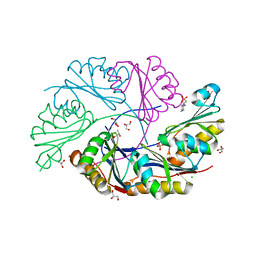 | | Ethanolamine Utilization Microcompartment Shell Subunit, EutS | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Ethanolamine utilization protein eutS, ... | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2009-07-10 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanisms of a Protein-Based Organelle in Escherichia coli.
Science, 327, 2010
|
|
3I87
 
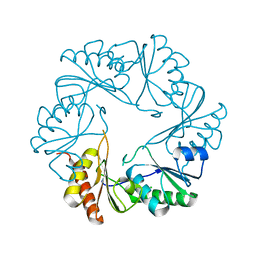 | |
3I71
 
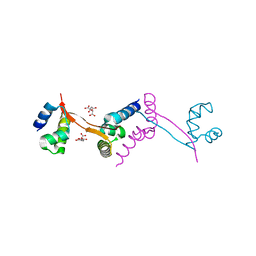 | |
3I82
 
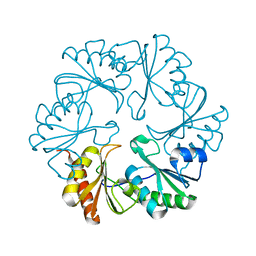 | |
3MLI
 
 | | 2ouf-ds, a disulfide-linked dimer of Helicobacter pylori protein HP0242 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | King, N.P, Sawaya, M.R, Jacobitz, A.W, Yeates, T.O. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and folding of a designed knotted protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MLG
 
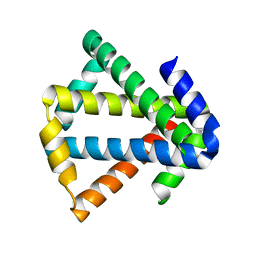 | | 2ouf-2x, a designed knotted protein | | Descriptor: | 2X chimera of Helicobacter pylori protein HP0242 | | Authors: | King, N.P, Sawaya, M.R, Jacobitz, A.W, Yeates, T.O. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and folding of a designed knotted protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5HRY
 
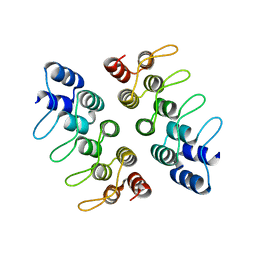 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HS0
 
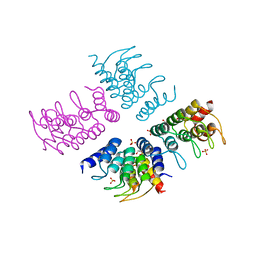 | | Computationally Designed Cyclic Tetramer ank1C4_7 | | Descriptor: | Ankyrin domain protein ank1C4_7, GLYCEROL, SULFATE ION | | Authors: | McNamara, D.E, Cascio, D, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HRZ
 
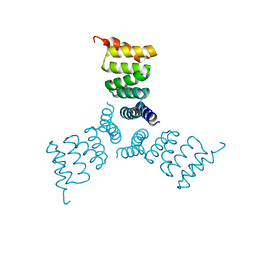 | | Computationally Designed Trimer 1na0C3_3 | | Descriptor: | TPR domain protein 1na0C3_3 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
4W6S
 
 | | Crystal Structure of Full-Length Split GFP Mutant K126C Disulfide Dimer, P 43 21 2 Space Group | | Descriptor: | GLYCEROL, PHOSPHATE ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W75
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 1 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.473 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7A
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 4 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6A
 
 | |
