8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z90
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z98
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8N
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z97
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8J
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z9R
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
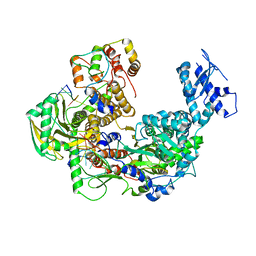 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8X
 
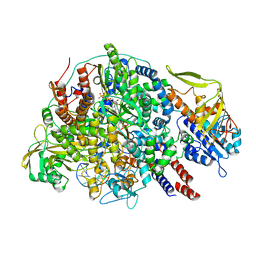 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
7OOC
 
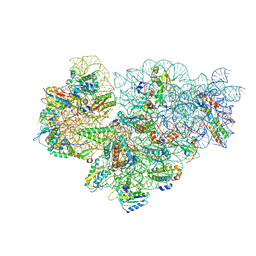 | | Mycoplasma pneumoniae 30S subunit of ribosomes in chloramphenicol-treated cells | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7OOD
 
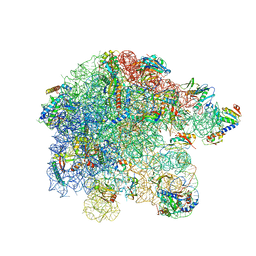 | | Mycoplasma pneumoniae 50S subunit of ribosomes in chloramphenicol-treated cells | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAI
 
 | | 70S ribosome with P-site tRNA in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAK
 
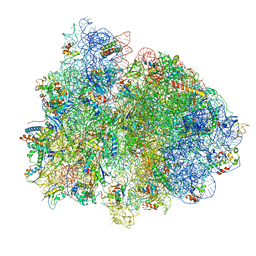 | | 70S ribosome with EF-Tu-tRNA and P-site tRNA in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7V6W
 
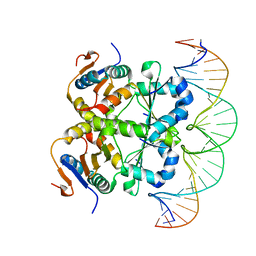 | | Crystal structure of heterohexameric Sa2YoeB-Sa2YefM complex bound to 26bp-DNA | | Descriptor: | Antitoxin, DNA (25-MER), DNA (26-MER), ... | | Authors: | Xue, L, Khan, M.H, Yue, J. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The two paralogous copies of the YoeB-YefM toxin-antitoxin module in Staphylococcus aureus differ in DNA binding and recognition patterns.
J.Biol.Chem., 298, 2022
|
|
7V5Y
 
 | |
7PAS
 
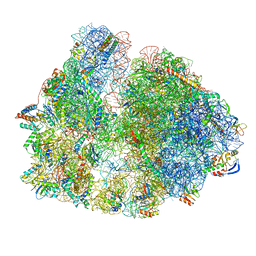 | | 70S ribosome with P/E-site tRNA in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAM
 
 | | 70S ribosome with A*- and P/E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAU
 
 | | free 50S in complex with ribosome recycling factor in untreated Mycoplasma pneumoniae cells | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7P6Z
 
 | | Mycoplasma pneumoniae 70S ribosome in untreated cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-18 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAL
 
 | | 70S ribosome with A- and P-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAH
 
 | | 70S ribosome with P- and E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7V5Z
 
 | |
7PAR
 
 | | 70S ribosome with EF-G, ap/P- and pe/E-site tRNAs in Mycoplasma pneumoniae cells | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
7PAT
 
 | | free 50S in untreated Mycoplasma pneumoniae cells | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Xue, L, Lenz, S, Rappsilber, J, Mahamid, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Visualizing translation dynamics at atomic detail inside a bacterial cell.
Nature, 610, 2022
|
|
