1IZB
 
 | |
1IZA
 
 | |
4CFF
 
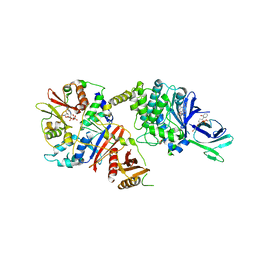 | | Structure of full length human AMPK in complex with a small molecule activator, a thienopyridone derivative (A-769662) | | Descriptor: | 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.924 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
4CFE
 
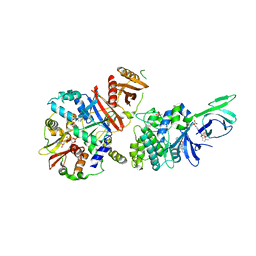 | | Structure of full length human AMPK in complex with a small molecule activator, a benzimidazole derivative (991) | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
1O9K
 
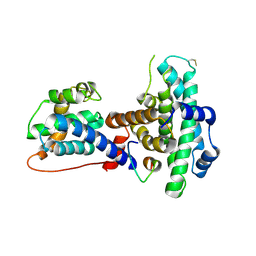 | | Crystal structure of the retinoblastoma tumour suppressor protein bound to E2F peptide | | Descriptor: | RETINOBLASTOMA-ASSOCIATED PROTEIN, TRANSCRIPTION FACTOR E2F1 | | Authors: | Xiao, B, Spencer, J, Clements, A, Ali-Khan, N, Mittnacht, S, Broceno, C, Burghammer, M, Perrakis, A, Marmorstein, R, Gamblin, S.J. | | Deposit date: | 2002-12-16 | | Release date: | 2003-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Retinoblastoma Tumor Suppressor Protein Bound to E2F and the Molecular Basis of its Regulation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1O9S
 
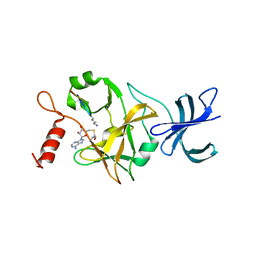 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
2V9J
 
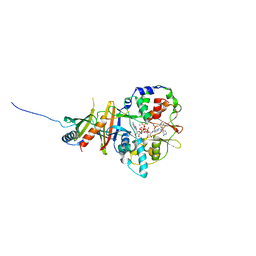 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V92
 
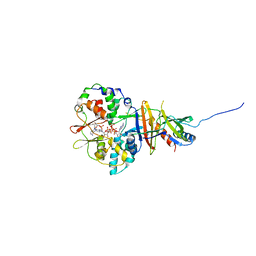 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V8Q
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
1EQM
 
 | | CRYSTAL STRUCTURE OF BINARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE WITH ADENOSINE-5'-DIPHOSPHATE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiao, B, Blaszczyk, J, Ji, X. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual conformational changes in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by X-ray crystallography and NMR.
J.Biol.Chem., 276, 2001
|
|
3QL9
 
 | |
3QLA
 
 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3BR8
 
 | |
2B5X
 
 | |
2B5Y
 
 | |
2AI6
 
 | |
1KKG
 
 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
2B8F
 
 | |
2B0G
 
 | |
2B8G
 
 | |
2AYM
 
 | |
1Z6H
 
 | | Solution Structure of Bacillus subtilis BLAP biotinylated-form | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Biotin/Lipoyl Attachment Protein | | Authors: | Cui, G, Xia, B. | | Deposit date: | 2005-03-22 | | Release date: | 2006-03-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Identification and solution structures of a single domain biotin/lipoyl attachment protein from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
1Z7R
 
 | | Solution Structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
1Z7P
 
 | | Solution structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
1J7C
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E95K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
