2JZE
 
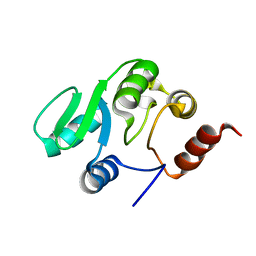 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2KLA
 
 | | NMR STRUCTURE OF A PUTATIVE DINITROGENASE (MJ0327) FROM METHANOCOCCUS JANNASCHII | | Descriptor: | Uncharacterized protein MJ0327 | | Authors: | Jaudzems, K, Mohanty, B, Geralt, M, Serrano, P, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_247299.1: comparison with the crystal structure.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KZF
 
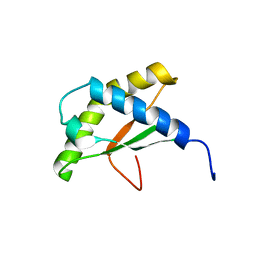 | | Solution NMR structure of the thermotoga maritima protein TM0855 a putative ribosome binding factor A | | Descriptor: | Ribosome-binding factor A | | Authors: | Serrano, P, Jaudzems, K, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the thermotoga maritima protein TM0855
To be Published
|
|
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
2HSX
 
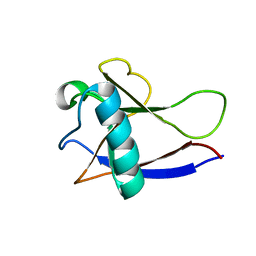 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-07-24 | | Release date: | 2007-02-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2KQV
 
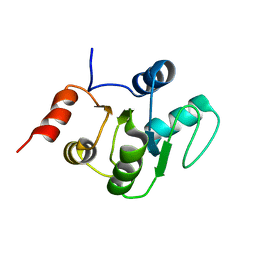 | |
2KAF
 
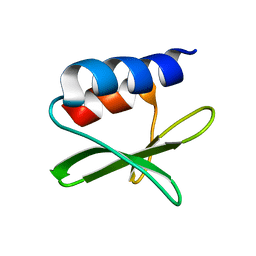 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
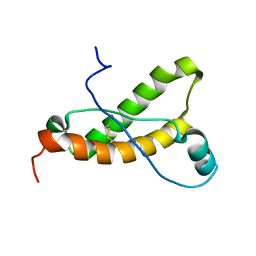
2KFM
 
 | |
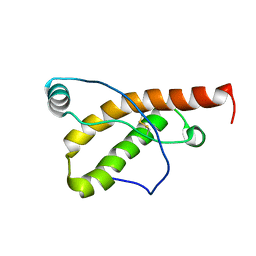
2KFL
 
 | |
2KFO
 
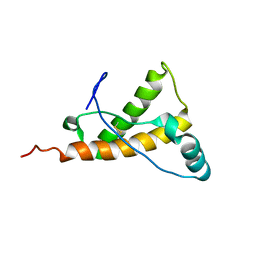 | |
2KU4
 
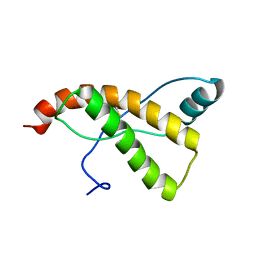 | | Horse prion protein | | Descriptor: | Major prion protein | | Authors: | Perez, D.R, Damberger, F.F, Wuthrich, K. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein.
J.Mol.Biol., 400, 2010
|
|
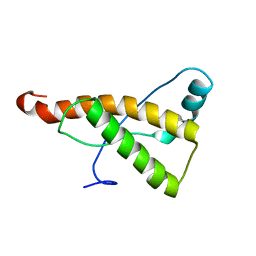
2KU5
 
 | |
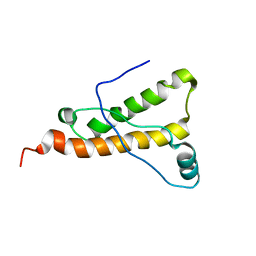
2KU6
 
 | |
2L39
 
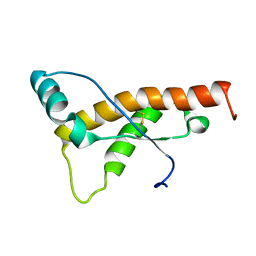 | | Mouse prion protein fragment 121-231 AT 37 C | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L1D
 
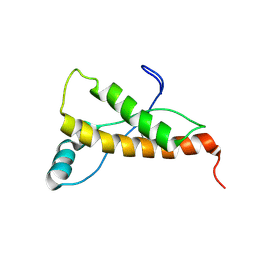 | | Mouse prion protein (121-231) containing the substitution Y169G | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2011-11-09 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L1E
 
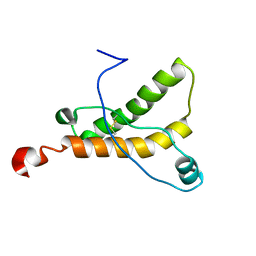 | | Mouse prion protein (121-231) containing the substitution F175A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2012-10-24 | | Method: | SOLUTION NMR | | Cite: | Prion Protein mPrP[F175A](121-231): Structure and Stability in Solution.
J.Mol.Biol., 423, 2012
|
|
2L1H
 
 | |
2L1K
 
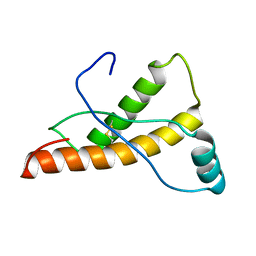 | | Mouse prion protein (121-231) containing the substitutions Y169A, Y225A, and Y226A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2012-02-01 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational exchange in the cellular form of prion proteins
To be Published
|
|
2L40
 
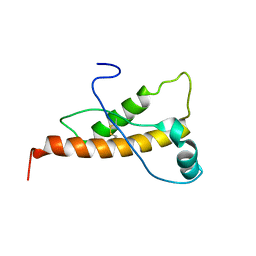 | | Mouse prion protein (121-231) containing the substitution Y169A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LFB
 
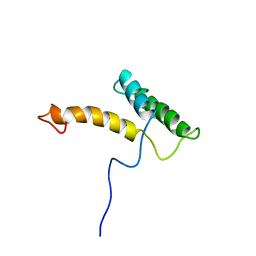 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | LFB1/HNF1 TRANSCRIPTION FACTOR | | Authors: | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | Deposit date: | 1996-12-12 | | Release date: | 1997-03-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
2K56
 
 | |
2K5O
 
 | |
2KL4
 
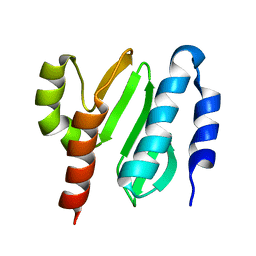 | | NMR structure of the protein NB7804A | | Descriptor: | BH2032 protein | | Authors: | Mohanty, B, Geralt, M, Augustyniak, W, Serrano, P, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein NB7804A from Bacillus Halodurans
To be Published
|
|
2K87
 
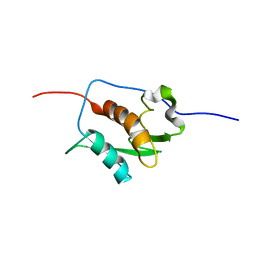 | | NMR STRUCTURE OF A PUTATIVE RNA BINDING PROTEIN (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | Non-structural protein 3 of Replicase polyprotein 1a | | Authors: | Serrano, P, Wuthrich, K, Johnson, M.A, Chatterjee, A, Wilson, I, Pedrini, B.F, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3.
J.Virol., 83, 2009
|
|
