1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN | | Authors: | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
4AIT
 
 | |
3EGF
 
 | |
2GDT
 
 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | Descriptor: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | Authors: | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-03-17 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
3AIT
 
 | |
3CYS
 
 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF THE CYCLOPHILIN A-CYCLOSPORIN A COMPLEX | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Spitzfaden, C, Braun, W, Wider, G, Widmer, H, Wuthrich, K. | | Deposit date: | 1994-02-28 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-01 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR Solution Structure of the Cyclophilin A-Cyclosporin a Complex.
J.Biomol.NMR, 4, 1994
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GA7
 
 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2RNK
 
 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1FTZ
 
 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
2BUS
 
 | |
2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1EGF
 
 | |
1HIC
 
 | |
1SAN
 
 | |
1MRB
 
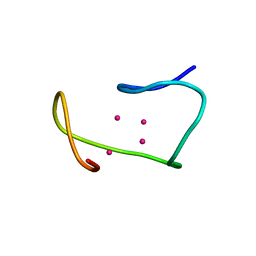 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
1HOM
 
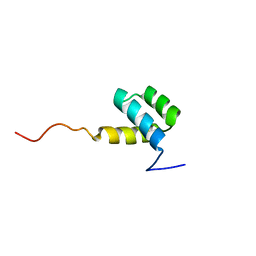 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Qian, Y.-Q, Billeter, M, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
2CCX
 
 | |
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1HD6
 
 | | PHEROMONE ER-22, NMR | | Descriptor: | PHEROMONE ER-22 | | Authors: | Luginbuhl, P, Liu, A, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 2000-11-09 | | Release date: | 2000-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Pheromone Er-22 from Euplotes Raikovi
J.Biomol.NMR, 19, 2001
|
|
1GRX
 
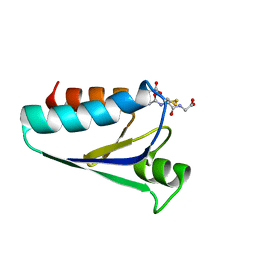 | | STRUCTURE OF E. COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN, GLUTATHIONE | | Authors: | Bushweller, J.H, Billeter, M, Holmgren, L.A, Wuthrich, K. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1QND
 
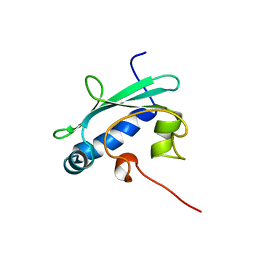 | | STEROL CARRIER PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Lopez-Garcia, F, Szyperski, T, Dyer, J.H, Choinowski, T, Seedorf, U, Hauser, H, Wuthrich, K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Sterol Carrier Protein-2: Implications for the Biological Role
J.Mol.Biol., 295, 2000
|
|
1MRT
 
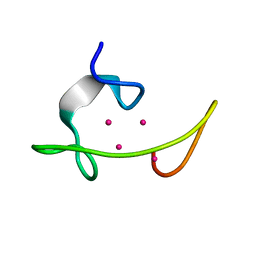 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
1ORM
 
 | |
1LG4
 
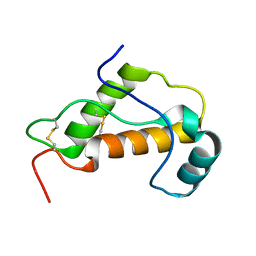 | |
