5AP8
 
 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | Descriptor: | TSR3 | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
5JP4
 
 | |
7PMQ
 
 | |
7PMM
 
 | |
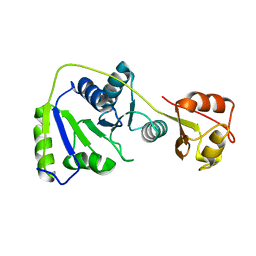
7BBB
 
 | |
5APG
 
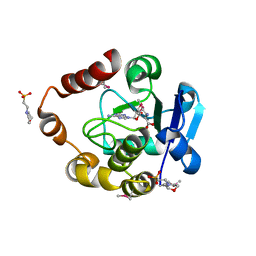 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
7PLI
 
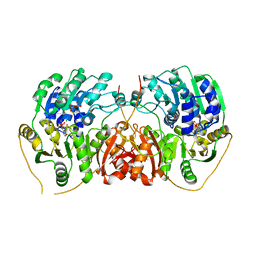 | | DEAD-box helicase DbpA bound to single stranded RNA and ADP/BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DbpA, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wurm, J.P. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA-duplex unwinding by the DEAD-box helicase DbpA.
Rna, 29, 2023
|
|
2MZJ
 
 | |
2N0J
 
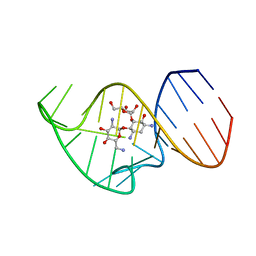 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | Descriptor: | RIBOSTAMYCIN, RNA_(27-MER) | | Authors: | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | Deposit date: | 2015-03-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7PVM
 
 | |
2LCQ
 
 | | Solution structure of the endonuclease Nob1 from P.horikoshii | | Descriptor: | Putative toxin VapC6, ZINC ION | | Authors: | Veith, T, Martin, R, Wurm, J.P, Weis, B, Duchardt-Ferner, E, Safferthal, C, Hennig, R, Mirus, O, Bohnsack, M.T, Woehnert, J, Schleiff, E. | | Deposit date: | 2011-05-05 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the archaeal endonuclease Nob1.
Nucleic Acids Res., 40, 2012
|
|
