6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | 登録日 | 2019-03-26 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | 登録日 | 2020-10-16 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D3U
 
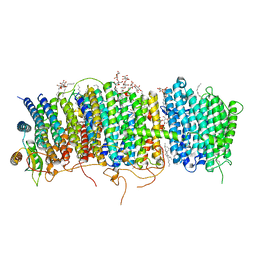 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | 分子名称: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | 著者 | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | 登録日 | 2020-09-21 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W5S
 
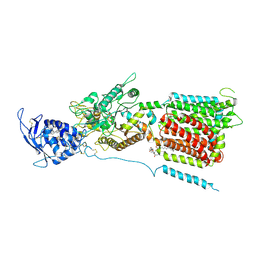 | | NPC1 structure in GDN micelles at pH 8.0 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5R
 
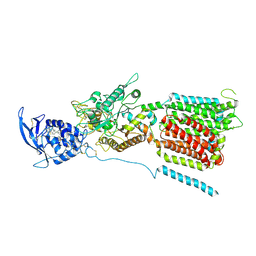 | | NPC1 structure in Nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5V
 
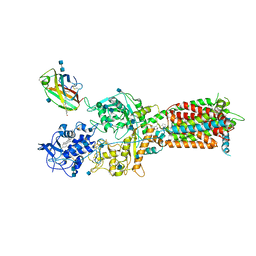 | | NPC1-NPC2 complex structure at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5T
 
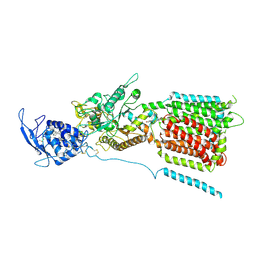 | | NPC1 structure in GDN micelles at pH 5.5, conformation a | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5U
 
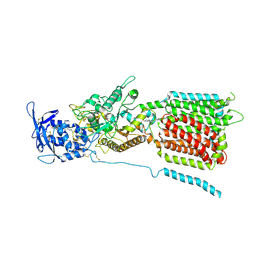 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
