6QE8
 
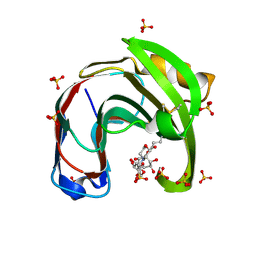 | | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-5-[(2~{S},3~{R},4~{S},5~{R})-3,4,5-tris(oxidanyl)oxan-2-yl]oxycyclohexane-1,2,3,4-tetrol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Wu, L, Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-01-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6GDT
 
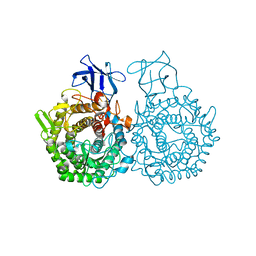 | |
5NPE
 
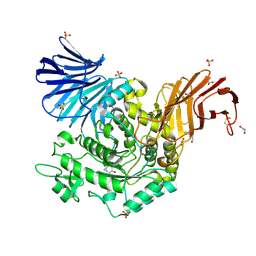 | | Crystal Structure of cjAgd31B (alpha-transglucosylase from Glycoside Hydrolase Family 31) in complex with beta Cyclophellitol Aziridine probe KY358 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5NPC
 
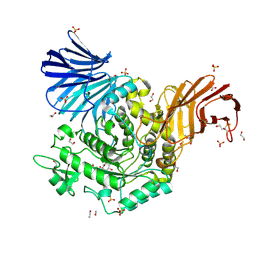 | | Crystal Structure of D412N nucleophile mutant cjAgd31B (alpha-transglucosylase from Glycoside Hydrolase Family 31) in complex with unreacted alpha Cyclophellitol Cyclosulfate probe ME647 | | Descriptor: | (3~{a}~{R},4~{R},5~{S},6~{R},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5NPB
 
 | |
5NPD
 
 | | Crystal Structure of D412N nucleophile mutant cjAgd31B (alpha-transglucosylase from Glycoside Hydrolase Family 31) in complex with alpha Cyclophellitol Aziridine probe CF021 | | Descriptor: | (1~{S},2~{S},3~{S},4~{R},5~{R},6~{S})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5NPF
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with beta Cyclophellitol Cyclosulfate probe ME594 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucosylceramidase, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5O0S
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
7P4C
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with noncovalent Cyclophellitol Sulfamidate probe KK131 | | Descriptor: | (3aR,4S,5S,6R,7R,7aS)-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3a,4,5,6,7,7a-hexahydro-3H-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7P4D
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with covalent Cyclophellitol Sulfamidate probe KK130 | | Descriptor: | 1,2-ETHANEDIOL, OXALATE ION, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7PR7
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VL166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PR8
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor GR109 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PRT
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor CB678 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-alpha-D-arabino-hexopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, ... | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PRB
 
 | | Crystal structure of Burkholderia pseudomallei heparanase in complex with covalent inhibitor GR109 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Glyco_hydro_44 domain-containing protein | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PR6
 
 | | Crystal structure of E. coli beta-glucuronidase in complex with covalent inhibitor ME727 | | Descriptor: | (2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Beta-glucuronidase | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PR9
 
 | | Crystal structure of Burkholderia pseudomallei heparanase in complex with covalent inhibitor VL166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-(2R,3S,5R,6R)-2,3,4,5,6-pentakis(oxidanyl)cyclohexane-1-carboxylic acid, Glyco_hydro_44 domain-containing protein | | Authors: | Wu, L, Armstrong, Z, Davies, G.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Mechanism-based heparanase inhibitors reduce cancer metastasis in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6YJS
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJQ
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A,Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJT
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJV
 
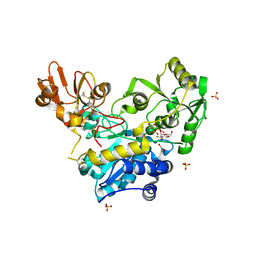 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP-2-deoxy-2-fluoroglucose and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJU
 
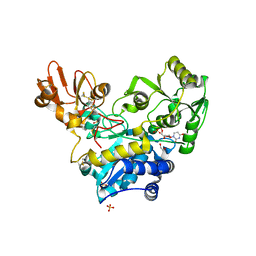 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJR
 
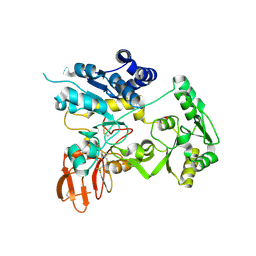 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
4JCL
 
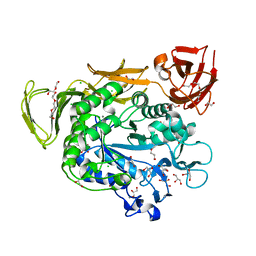 | | Crystal structure of Alpha-CGT from Paenibacillus macerans at 1.7 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J, Wu, J, Li, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Alpha-Cgt from Paenibacillus Macerans at 1.7 Angstrom Resolution
To be Published
|
|
8OHQ
 
 | | Crystal structure of human heparanase in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Davies, G.J, Overkleeft, H.S, Armstrong, Z, Yurong, C. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
4JCM
 
 | | Crystal structure of Gamma-CGTASE from Alkalophilic bacillus clarkii at 1.65 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Yang, D, Zhou, J, Wu, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Gamma-Cgtase from Alkalophilic Bacillus Clarkii at 1.65 Angstrom Resolution.
To be Published
|
|
